Trace amine associated receptor 1: predicted effects of single nucleotide variants on structure-function in geographically diverse populations
- PMID: 38863077
- PMCID: PMC11165750
- DOI: 10.1186/s40246-024-00620-w
Trace amine associated receptor 1: predicted effects of single nucleotide variants on structure-function in geographically diverse populations
Abstract
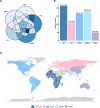
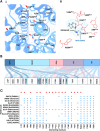
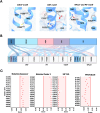
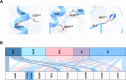
Trace Amine Associated Receptor 1 (TAAR1) is a novel pharmaceutical target under investigation for the treatment of several neuropsychiatric conditions. TAAR1 single nucleotide variants (SNV) have been found in patients with schizophrenia and metabolic disorders. However, the frequency of variants in geographically diverse populations and the functional effects of such variants are unknown. In this study, we aimed to characterise the distribution of TAAR1 SNVs in five different WHO regions using the Database of Genotypes and Phenotypes (dbGaP) and conducted a critical computational analysis using available TAAR1 structural data to identify SNVs affecting ligand binding and/or functional regions. Our analysis shows 19 orthosteric, 9 signalling and 16 micro-switch SNVs hypothesised to critically influence the agonist induced TAAR1 activation. These SNVs may non-proportionally influence populations from discrete regions and differentially influence the activity of TAAR1-targeting therapeutics in genetically and geographically diverse populations. Notably, our dataset presented with orthosteric SNVs D1033.32N (found only in the South-East Asian Region and Western Pacific Region) and T1945.42A (found only in South-East Asian Region), and 2 signalling SNVs (V1253.54A/T2526.36A, found in African Region and commonly, respectively), all of which have previously demonstrated to influence ligand induced functions of TAAR1. Furthermore, bioinformatics analysis using SIFT4G, MutationTaster 2, PROVEAN and MutationAssessor predicted all 16 micro-switch SNVs are damaging and may further influence the agonist activation of TAAR1, thereby possibly impacting upon clinical outcomes. Understanding the genetic basis of TAAR1 function and the impact of common mutations within clinical populations is important for the safe and effective utilisation of novel and existing pharmacotherapies.
Keywords: Agonists; Aminergic receptors; Amphetamines; Demographic; Genetic variants; Neuropsychiatric disorders; Neurotransmitters; Ralmitaront; Trace amines; Ulotaront.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Unlocking the secrets of trace amine-associated receptor 1 agonists: new horizon in neuropsychiatric treatment.Front Psychiatry. 2024 Oct 31;15:1464550. doi: 10.3389/fpsyt.2024.1464550. eCollection 2024. Front Psychiatry. 2024. PMID: 39553890 Free PMC article. Review.
-
Trace amine-associated receptor 1 (TAAR1) agonism for psychosis: a living systematic review and meta-analysis of human and non-human data.Wellcome Open Res. 2024 Apr 11;9:182. doi: 10.12688/wellcomeopenres.21302.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39036710 Free PMC article.
-
In Vitro Comparison of Ulotaront (SEP-363856) and Ralmitaront (RO6889450): Two TAAR1 Agonist Candidate Antipsychotics.Int J Neuropsychopharmacol. 2023 Sep 25;26(9):599-606. doi: 10.1093/ijnp/pyad049. Int J Neuropsychopharmacol. 2023. PMID: 37549917 Free PMC article.
-
Therapeutic Potential of TAAR1 Agonists in Schizophrenia: Evidence from Preclinical Models and Clinical Studies.Int J Mol Sci. 2021 Dec 7;22(24):13185. doi: 10.3390/ijms222413185. Int J Mol Sci. 2021. PMID: 34947997 Free PMC article. Review.
-
Differential modulation of Beta-adrenergic receptor signaling by trace amine-associated receptor 1 agonists.PLoS One. 2011;6(10):e27073. doi: 10.1371/journal.pone.0027073. Epub 2011 Oct 31. PLoS One. 2011. PMID: 22073124 Free PMC article.
Cited by
-
Unlocking the secrets of trace amine-associated receptor 1 agonists: new horizon in neuropsychiatric treatment.Front Psychiatry. 2024 Oct 31;15:1464550. doi: 10.3389/fpsyt.2024.1464550. eCollection 2024. Front Psychiatry. 2024. PMID: 39553890 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

