Development and validation of CYP26A1 inhibition assay for high-throughput screening
- PMID: 38863121
- PMCID: PMC11338008
- DOI: 10.1002/biot.202300659
Development and validation of CYP26A1 inhibition assay for high-throughput screening
Abstract
All-trans retinoic acid (atRA) is an endogenous ligand of the retinoic acid receptors, which heterodimerize with retinoid X receptors. AtRA is generated in tissues from vitamin A (retinol) metabolism to form a paracrine signal and is locally degraded by cytochrome P450 family 26 (CYP26) enzymes. The CYP26 family consists of three subtypes: A1, B1, and C1, which are differentially expressed during development. This study aims to develop and validate a high throughput screening assay to identify CYP26A1 inhibitors in a cell-free system using a luminescent P450-Glo assay technology. The assay performed well with a signal to background ratio of 25.7, a coefficient of variation of 8.9%, and a Z-factor of 0.7. To validate the assay, we tested a subset of 39 compounds that included known CYP26 inhibitors and retinoids, as well as positive and negative control compounds selected from the literature and/or the ToxCast/Tox21 portfolio. Known CYP26A1 inhibitors were confirmed, and predicted CYP26A1 inhibitors, such as chlorothalonil, prochloraz, and SSR126768, were identified, demonstrating the reliability and robustness of the assay. Given the general importance of atRA as a morphogenetic signal and the localized expression of Cyp26a1 in embryonic tissues, a validated CYP26A1 assay has important implications for evaluating the potential developmental toxicity of chemicals.
Keywords: CYP26; all‐trans retinoic acid (atRA); cytochrome P450 (CYP); retinoic acid receptor (RAR).
Published 2024. This article is a U.S. Government work and is in the public domain in the USA. Biotechnology Journal published by Wiley‐VCH GmbH.
Conflict of interest statement
CONFLICT OF INTEREST STATEMENT
The authors declare no commercial or financial conflict of interest.
Figures



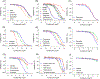
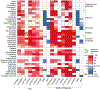
References
-
- Bastien J, & Rochette-Egly C. (2004). Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene, 328, 1–16. - PubMed
-
- Marill J, Idres N, Capron CC, Nguyen E, & Chabot GG (2003). Retinoic acid metabolism and mechanism of action: A review. Current Drug Metabolism, 4(1), 1–10. - PubMed
-
- McSorley LC, & Daly AK (2000). Identification of human cytochrome P450 isoforms that contribute to all-trans-retinoic acid 4-hydroxylation. Biochemical Pharmacology, 60(4), 517–526. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

