Critical cis-parameters influence STructure assisted RNA translation (START) initiation on non-AUG codons in eukaryotes
- PMID: 38863530
- PMCID: PMC11165317
- DOI: 10.1093/nargab/lqae065
Critical cis-parameters influence STructure assisted RNA translation (START) initiation on non-AUG codons in eukaryotes
Abstract
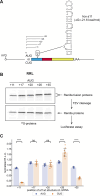
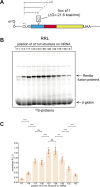
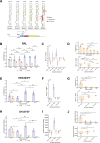
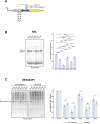
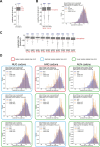
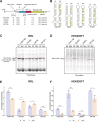
In eukaryotes, translation initiation is a highly regulated process, which combines cis-regulatory sequences located on the messenger RNA along with trans-acting factors like eukaryotic initiation factors (eIF). One critical step of translation initiation is the start codon recognition by the scanning 43S particle, which leads to ribosome assembly and protein synthesis. In this study, we investigated the involvement of secondary structures downstream the initiation codon in the so-called START (STructure-Assisted RNA translation) mechanism on AUG and non-AUG translation initiation. The results demonstrate that downstream secondary structures can efficiently promote non-AUG translation initiation if they are sufficiently stable to stall a scanning 43S particle and if they are located at an optimal distance from non-AUG codons to stabilize the codon-anticodon base pairing in the P site. The required stability of the downstream structure for efficient translation initiation varies in distinct cell types. We extended this study to genome-wide analysis of functionally characterized alternative translation initiation sites in Homo sapiens. This analysis revealed that about 25% of these sites have an optimally located downstream secondary structure of adequate stability which could elicit START, regardless of the start codon. We validated the impact of these structures on translation initiation for several selected uORFs.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures

Similar articles
-
AUG_hairpin: prediction of a downstream secondary structure influencing the recognition of a translation start site.BMC Bioinformatics. 2007 Aug 30;8:318. doi: 10.1186/1471-2105-8-318. BMC Bioinformatics. 2007. PMID: 17760957 Free PMC article.
-
The interplay between cis- and trans-acting factors drives selective mRNA translation initiation in eukaryotes.Biochimie. 2024 Feb;217:20-30. doi: 10.1016/j.biochi.2023.09.017. Epub 2023 Sep 21. Biochimie. 2024. PMID: 37741547 Review.
-
Alternative Translation Initiation of a Haloarchaeal Serine Protease Transcript Containing Two In-Frame Start Codons.J Bacteriol. 2016 Jun 13;198(13):1892-901. doi: 10.1128/JB.00202-16. Print 2016 Jul 1. J Bacteriol. 2016. PMID: 27137502 Free PMC article.
-
Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation.RNA. 2009 Jan;15(1):138-52. doi: 10.1261/rna.1318509. Epub 2008 Nov 24. RNA. 2009. PMID: 19029312 Free PMC article.
-
Initiation of translation in prokaryotes and eukaryotes.Gene. 1999 Jul 8;234(2):187-208. doi: 10.1016/s0378-1119(99)00210-3. Gene. 1999. PMID: 10395892 Review.
Cited by
-
Impact of codon optimization on vip3Aa11 gene expression and insecticidal efficacy in maize.Front Plant Sci. 2025 May 13;16:1579465. doi: 10.3389/fpls.2025.1579465. eCollection 2025. Front Plant Sci. 2025. PMID: 40433150 Free PMC article.
References
-
- Tidu A., Martin F. The interplay between cis- and trans-acting factors drives selective mRNA translation initiation in eukaryotes. Biochimie. 2023; 217:20–30. - PubMed
-
- Rogers G.W., Richter N.J., Lima W.F., Merrick W.C. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001; 276:30914–30922. - PubMed
-
- Passmore L.A., Schmeing T.M., Maag D., Applefield D.J., Acker M.G., Algire M.A., Lorsch J.R., Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007; 26:41–50. - PubMed
LinkOut - more resources
Full Text Sources

