Insight into chromatin compaction and spatial organization in rice interphase nuclei
- PMID: 38863533
- PMCID: PMC11165205
- DOI: 10.3389/fpls.2024.1358760
Insight into chromatin compaction and spatial organization in rice interphase nuclei
Abstract
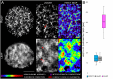
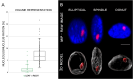
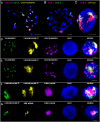
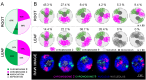
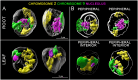
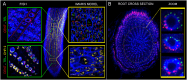
Chromatin organization and its interactions are essential for biological processes, such as DNA repair, transcription, and DNA replication. Detailed cytogenetics data on chromatin conformation, and the arrangement and mutual positioning of chromosome territories in interphase nuclei are still widely missing in plants. In this study, level of chromatin condensation in interphase nuclei of rice (Oryza sativa) and the distribution of chromosome territories (CTs) were analyzed. Super-resolution, stimulated emission depletion (STED) microscopy showed different levels of chromatin condensation in leaf and root interphase nuclei. 3D immuno-FISH experiments with painting probes specific to chromosomes 9 and 2 were conducted to investigate their spatial distribution in root and leaf nuclei. Six different configurations of chromosome territories, including their complete association, weak association, and complete separation, were observed in root meristematic nuclei, and four configurations were observed in leaf nuclei. The volume of CTs and frequency of their association varied between the tissue types. The frequency of association of CTs specific to chromosome 9, containing NOR region, is also affected by the activity of the 45S rDNA locus. Our data suggested that the arrangement of chromosomes in the nucleus is connected with the position and the size of the nucleolus.
Keywords: 3D immuno-FISH; chromosome painting; chromosome territory; microscopy; rice; spatial organization.
Copyright © 2024 Doležalová, Beránková, Koláčková and Hřibová.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
The arrangement of Brachypodium distachyon chromosomes in interphase nuclei.J Exp Bot. 2016 Oct;67(18):5571-5583. doi: 10.1093/jxb/erw325. Epub 2016 Sep 1. J Exp Bot. 2016. PMID: 27588463 Free PMC article.
-
Interphase chromatin organisation in Arabidopsis nuclei: constraints versus randomness.Chromosoma. 2012 Aug;121(4):369-87. doi: 10.1007/s00412-012-0367-8. Epub 2012 Apr 4. Chromosoma. 2012. PMID: 22476443
-
Simulation of Different Three-Dimensional Models of Whole Interphase Nuclei Compared to Experiments - A Consistent Scale-Bridging Simulation Framework for Genome Organization.Results Probl Cell Differ. 2022;70:495-549. doi: 10.1007/978-3-031-06573-6_18. Results Probl Cell Differ. 2022. PMID: 36348120
-
Simulation of different three-dimensional polymer models of interphase chromosomes compared to experiments-an evaluation and review framework of the 3D genome organization.Semin Cell Dev Biol. 2019 Jun;90:19-42. doi: 10.1016/j.semcdb.2018.07.012. Epub 2018 Aug 24. Semin Cell Dev Biol. 2019. PMID: 30125668 Review.
-
Organization and dynamics of plant interphase chromosomes.Trends Plant Sci. 2011 May;16(5):273-81. doi: 10.1016/j.tplants.2011.02.002. Epub 2011 Mar 9. Trends Plant Sci. 2011. PMID: 21393049 Review.
Cited by
-
Thriving or Withering? Plant Molecular Cytogenetics in the First Quarter of the 21st Century.Int J Mol Sci. 2025 Jul 21;26(14):7013. doi: 10.3390/ijms26147013. Int J Mol Sci. 2025. PMID: 40725259 Free PMC article. Review.
References
-
- Abbe E. (1873). Beitrage zur Theorie des Mikroskops und der mikroskopischen Wahrmehmung. Archiv für mikroskopische Anatomie (in German). 9, 413–420. doi: 10.1007/BF02956173 - DOI
-
- Anamthawat-Jónsson K., Heslop-Harrison J. S. (1990). Centromeres, telomeres and chromatin in the interphase nucleus of cereals. Caryologia 43, 205–213. doi: 10.1080/00087114.1990.10796999 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

