Genome-wide identification of bZIP transcription factors and their expression analysis in Platycodon grandiflorus under abiotic stress
- PMID: 38863542
- PMCID: PMC11165138
- DOI: 10.3389/fpls.2024.1403220
Genome-wide identification of bZIP transcription factors and their expression analysis in Platycodon grandiflorus under abiotic stress
Abstract
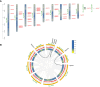
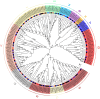
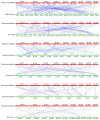
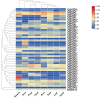
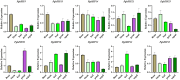
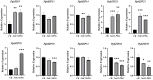
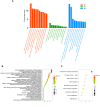
The Basic Leucine Zipper (bZIP) transcription factors (TFs) family is among of the largest and most diverse gene families found in plant species, and members of the bZIP TFs family perform important functions in plant developmental processes and stress response. To date, bZIP genes in Platycodon grandiflorus have not been characterized. In this work, a number of 47 PgbZIP genes were identified from the genome of P. grandiflorus, divided into 11 subfamilies. The distribution of these PgbZIP genes on the chromosome and gene replication events were analyzed. The motif, gene structure, cis-elements, and collinearity relationships of the PgbZIP genes were simultaneously analyzed. In addition, gene expression pattern analysis identified ten candidate genes involved in the developmental process of different tissue parts of P. grandiflorus. Among them, Four genes (PgbZIP5, PgbZIP21, PgbZIP25 and PgbZIP28) responded to drought and salt stress, which may have potential biological roles in P. grandiflorus development under salt and drought stress. Four hub genes (PgbZIP13, PgbZIP30, PgbZIP32 and PgbZIP45) mined in correlation network analysis, suggesting that these PgbZIP genes may form a regulatory network with other transcription factors to participate in regulating the growth and development of P. grandiflorus. This study provides new insights regarding the understanding of the comprehensive characterization of the PgbZIP TFs for further exploration of the functions of growth and developmental regulation in P. grandiflorus and the mechanisms for coping with abiotic stress response.
Keywords: Platycodon grandiflorus; abiotic stress; bZIP transcription factor; evolutionary analyses; expression profiling.
Copyright © 2024 Wang, Wang, Cao, Liu, Kong, Wang, Ren, Fu and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Genome-wide analysis of bZIP transcription factors and their expression patterns in response to methyl jasmonate and low-temperature stresses in Platycodon grandiflorus.PeerJ. 2024 Apr 30;12:e17371. doi: 10.7717/peerj.17371. eCollection 2024. PeerJ. 2024. PMID: 38708338 Free PMC article.
-
Basic leucine zipper (bZIP) transcription factor genes and their responses to drought stress in ginseng, Panax ginseng C.A. Meyer.BMC Genomics. 2021 May 1;22(1):316. doi: 10.1186/s12864-021-07624-z. BMC Genomics. 2021. PMID: 33932982 Free PMC article.
-
"Genome-wide identification of bZIP gene family in Pearl millet and transcriptional profiling under abiotic stress, phytohormonal treatments; and functional characterization of PgbZIP9".Front Plant Sci. 2024 Feb 26;15:1352040. doi: 10.3389/fpls.2024.1352040. eCollection 2024. Front Plant Sci. 2024. PMID: 38469329 Free PMC article.
-
Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: A molecular model of a wheat bZIP factor and implications of its structure in function.Biochim Biophys Acta. 2016 Jan;1860(1 Pt A):46-56. doi: 10.1016/j.bbagen.2015.10.014. Epub 2015 Oct 20. Biochim Biophys Acta. 2016. PMID: 26493723 Review.
-
bZIP Transcription Factors: Structure, Modification, Abiotic Stress Responses and Application in Plant Improvement.Plants (Basel). 2024 Jul 25;13(15):2058. doi: 10.3390/plants13152058. Plants (Basel). 2024. PMID: 39124175 Free PMC article. Review.
Cited by
-
Unraveling TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR Transcription Factors in Safflower: A Blueprint for Stress Resilience and Metabolic Regulation.Molecules. 2025 Jan 10;30(2):254. doi: 10.3390/molecules30020254. Molecules. 2025. PMID: 39860123 Free PMC article.
-
Identification and functional characterization of JrbZIP40 in walnut reveals its role in salt and drought stress tolerance in transgenic Arabidopsis seedlings.BMC Plant Biol. 2025 Jul 2;25(1):807. doi: 10.1186/s12870-025-06928-6. BMC Plant Biol. 2025. PMID: 40604448 Free PMC article.
-
Application of proteomics in investigating the responses of plant to abiotic stresses.Planta. 2025 May 7;261(6):128. doi: 10.1007/s00425-025-04707-z. Planta. 2025. PMID: 40332605 Review.
References
Associated data
LinkOut - more resources
Full Text Sources

