Comprehensive analysis of the Spartina alterniflora WD40 gene family reveals the regulatory role of SaTTG1 in plant development
- PMID: 38863548
- PMCID: PMC11165199
- DOI: 10.3389/fpls.2024.1390461
Comprehensive analysis of the Spartina alterniflora WD40 gene family reveals the regulatory role of SaTTG1 in plant development
Abstract
Introduction: The WD40 gene family, prevalent in eukaryotes, assumes diverse roles in cellular processes. Spartina alterniflora, a halophyte with exceptional salt tolerance, flood tolerance, reproduction, and diffusion ability, offers great potential for industrial applications and crop breeding analysis. The exploration of growth and development-related genes in this species offers immense potential for enhancing crop yield and environmental adaptability, particularly in industrialized plantations. However, the understanding of their role in regulating plant growth and development remains limited.
Methods: In this study, we conducted a comprehensive analysis of WD40 genes in S. alterniflora at the whole-genome level, delving into their characteristics such as physicochemical properties, phylogenetic relationships, gene architecture, and expression patterns. Additionally, we cloned the TTG1 gene, a gene in plant growth and development across diverse species.
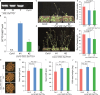
Results: We identified a total of 582 WD40 proteins in the S. alterniflora genome, exhibiting an uneven distribution across chromosomes. Through phylogenetic analysis, we categorized the 582 SaWD40 proteins into 12 distinct clades. Examining the duplication patterns of SaWD40 genes, we observed a predominant role of segmental duplication in their expansion. A substantial proportion of SaWD40 gene duplication pairs underwent purifying selection through evolution. To explore the functional aspects, we selected SaTTG1, a homolog of Arabidopsis TTG1, for overexpression in Arabidopsis. Subcellular localization analysis revealed that the SaTTG1 protein localized in the nucleus and plasma membrane, exhibiting transcriptional activation in yeast cells. The overexpression of SaTTG1 in Arabidopsis resulted in early flowering and increased seed size.
Discussion: These outcomes significantly contribute to our understanding of WD40 gene functions in halophyte species. The findings not only serve as a valuable foundation for further investigations into WD40 genes in halophyte but also offer insights into the molecular mechanisms governing plant development, offering potential avenues in molecular breeding.
Keywords: Spartina alterniflora; TTG1; WD40; flowering time; seed size.
Copyright © 2024 Yang, Chen, Geng, Gao, Chen and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer ZL declared a shared affiliation with the authors MY, SC, and HL to the handling editor at the time of review. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures




References
-
- Ben-Simhon Z., Judeinstein S., Nadler-Hassar T., Trainin T., Bar-Ya'akov I., Borochov-Neori H., et al. . (2011). A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 234, 865–881. doi: 10.1007/s00425-011-1438-4 - DOI - PubMed
LinkOut - more resources
Full Text Sources

