Dengue virus exploits autophagy vesicles and secretory pathways to promote transmission by human dendritic cells
- PMID: 38863700
- PMCID: PMC11165123
- DOI: 10.3389/fimmu.2024.1260439
Dengue virus exploits autophagy vesicles and secretory pathways to promote transmission by human dendritic cells
Abstract
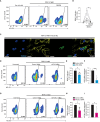
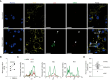
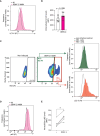
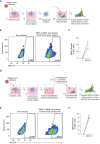
Dengue virus (DENV), transmitted by infected mosquitoes, is a major public health concern, with approximately half the world's population at risk for infection. Recent decades have increasing incidence of dengue-associated disease alongside growing frequency of outbreaks. Although promising progress has been made in anti-DENV immunizations, post-infection treatment remains limited to non-specific supportive treatments. Development of antiviral therapeutics is thus required to limit DENV dissemination in humans and to help control the severity of outbreaks. Dendritic cells (DCs) are amongst the first cells to encounter DENV upon injection into the human skin mucosa, and thereafter promote systemic viral dissemination to additional human target cells. Autophagy is a vesicle trafficking pathway involving the formation of cytosolic autophagosomes, and recent reports have highlighted the extensive manipulation of autophagy by flaviviruses, including DENV, for viral replication. However, the temporal profiling and function of autophagy activity in DENV infection and transmission by human primary DCs remains poorly understood. Herein, we demonstrate that mechanisms of autophagosome formation and extracellular vesicle (EV) release have a pro-viral role in DC-mediated DENV transmission. We show that DENV exploits early-stage canonical autophagy to establish infection in primary human DCs. DENV replication enhanced autophagosome formation in primary human DCs, and intrinsically-heightened autophagosome biogenesis correlated with relatively higher rates of DC susceptibility to DENV. Furthermore, our data suggest that viral replication intermediates co-localize with autophagosomes, while productive DENV infection introduces a block at the late degradative stages of autophagy in infected DCs but not in uninfected bystander cells. Notably, we identify for the first time that approximately one-fourth of DC-derived CD9/CD81/CD63+ EVs co-express canonical autophagy marker LC3, and demonstrate that DC-derived EV populations are an alternative, cell-free mechanism by which DCs promote DENV transmission to additional target sites. Taken together, our study highlights intersections between autophagy and secretory pathways during viral infection, and puts forward autophagosome accumulation and viral RNA-laden EVs as host determinants of DC-mediated DENV infection in humans. Host-directed therapeutics targeting autophagy and exocytosis pathways thus have potential to enhance DC-driven resistance to DENV acquisition and thereby limit viral dissemination by initial human target cells following mosquito-to-human transmission of DENV.
Keywords: autophagy; dendritic cells; dengue virus; extracellular vesicles; host-directed antivirals; secretory autophagy; viral evasion; viral transmission.
Copyright © 2024 Cloherty, Rader, Patel, Eisden, van Piggelen, Schreurs and Ribeiro.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- World Health Organization . Dengue and severe dengue factsheet(2022). Available at: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
-
- Dengue vaccine. WHO position paper, September 2018 - recommendations(2018). Available at: https://www.who.int/publications/i/item/who-wer9335-457-476. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

