Genomic and phenotypic characterization of 26 novel marine bacterial strains with relevant biogeochemical roles and widespread presence across the global ocean
- PMID: 38863746
- PMCID: PMC11165706
- DOI: 10.3389/fmicb.2024.1407904
Genomic and phenotypic characterization of 26 novel marine bacterial strains with relevant biogeochemical roles and widespread presence across the global ocean
Abstract
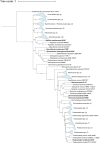
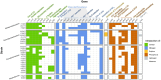
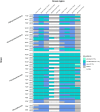
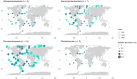
Prokaryotes dominate global oceans and shape biogeochemical cycles, yet most taxa remain uncultured and uncharacterized as of today. Here we present the characterization of 26 novel marine bacterial strains from a large isolate collection obtained from Blanes Bay (NW Mediterranean) microcosm experiments made in the four seasons. Morphological, cultural, biochemical, physiological, nutritional, genomic, and phylogenomic analyses were used to characterize and phylogenetically place the novel isolates. The strains represent 23 novel bacterial species and six novel genera: three novel species pertaining to class Alphaproteobacteria (families Rhodobacteraceae and Sphingomonadaceae), six novel species and three new genera from class Gammaproteobacteria (families Algiphilaceae, Salinispheraceae, and Alteromonadaceae), 13 novel species and three novel genera from class Bacteroidia (family Flavobacteriaceae), and one new species from class Rhodothermia (family Rubricoccaceae). The bacteria described here have potentially relevant roles in the cycles of carbon (e.g., carbon fixation or energy production via proteorhodopsin), nitrogen (e.g., denitrification or use of urea), sulfur (oxidation of sulfur compounds), phosphorus (acquisition and use of different forms of phosphorus and remodeling of membrane phospholipids), and hydrogen (oxidation of hydrogen to obtain energy). We mapped the genomes of the presented strains to the Tara Oceans metagenomes to reveal that these strains were globally distributed, with those of the family Flavobacteriaceae being the most widespread and abundant, while Rhodothermia being the rarest and most localized. While molecular-only approaches are also important, our study stresses the importance of culturing as a powerful tool to further understand the functioning of marine bacterial communities.
Keywords: biogeochemistry; biogeography; genomics; isolates; marine bacteria; phylogenomics; taxonomy.
Copyright © 2024 Rey-Velasco, Lucena, Belda, Gasol, Sánchez, Arahal and Pujalte.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be considered as a potential conflict of interest.
Figures



Similar articles
-
Diversity and distribution of marine heterotrophic bacteria from a large culture collection.BMC Microbiol. 2020 Jul 13;20(1):207. doi: 10.1186/s12866-020-01884-7. BMC Microbiol. 2020. PMID: 32660423 Free PMC article.
-
Genomic analysis and characterization of phages infecting the marine Roseobacter CHAB-I-5 lineage reveal a globally distributed and abundant phage genus.Front Microbiol. 2023 Apr 17;14:1164101. doi: 10.3389/fmicb.2023.1164101. eCollection 2023. Front Microbiol. 2023. PMID: 37138617 Free PMC article.
-
Expanding success in the isolation of abundant marine bacteria after reduction in grazing and viral pressure and increase in nutrient availability.Microbiol Spectr. 2023 Sep 25;11(5):e0089023. doi: 10.1128/spectrum.00890-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37747249 Free PMC article.
-
The Minderoo-Monaco Commission on Plastics and Human Health.Ann Glob Health. 2023 Mar 21;89(1):23. doi: 10.5334/aogh.4056. eCollection 2023. Ann Glob Health. 2023. PMID: 36969097 Free PMC article. Review.
-
Microbial ecology of ocean biogeochemistry: a community perspective.Science. 2008 May 23;320(5879):1043-5. doi: 10.1126/science.1153527. Science. 2008. PMID: 18497289 Review.
Cited by
-
Bringing the uncultivated microbial majority of freshwater ecosystems into culture.Nat Commun. 2025 Aug 26;16(1):7971. doi: 10.1038/s41467-025-63266-9. Nat Commun. 2025. PMID: 40858551 Free PMC article.
References
-
- Azam F., Fenchel T., Field J., Gray J., Meyer-Reil L., Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257 - DOI
LinkOut - more resources
Full Text Sources

