Identification of Key Disulfidptosis-Related Genes and Their Association with Gene Expression Subtypes in Crohn's Disease
- PMID: 38863903
- PMCID: PMC11166158
- DOI: 10.2147/JIR.S458951
Identification of Key Disulfidptosis-Related Genes and Their Association with Gene Expression Subtypes in Crohn's Disease
Abstract
Background: Crohn's disease (CD) is a persistent inflammatory condition that impacts the gastrointestinal system and is characterized by a multifaceted pathogenesis involving genetic, immune, and environmental components. This study primarily investigates the relationship between gene expression and immune cell infiltration in CD, focusing on disulfidptosis-a novel form of cell death caused by abnormal disulfide accumulation-and its impact on various immune cell populations. By identifying key disulfidptosis-related genes (DRGs) and exploring their association with distinct gene expression subtypes, this research aims to enhance our understanding of CD and potentially other autoimmune diseases.
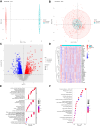
Methods: Gene expression data from intestinal biopsy samples were collected from both individuals with CD and healthy controls, and these data were retrieved from the GEO database. Through gene expression level comparisons, various differentially expressed genes (DEGs) were identified. Subsequently, Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed to reveal the biological processes and pathways linked to these DEGs. Later, immune cell infiltration was evaluated. Hub candidate DRGs were identified using machine learning algorithms. Validation of the expression of hub DRGs was carried out using quantitative real-time polymerase chain reaction. The hub DRGs were subjected to unsupervised hierarchical clustering to classify CD patients into subtypes. The characteristics of each subtype were then analyzed.
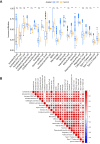
Results: Two hub DRGs (NDUFA11 and LRPPRC) were identified. NDUFA11 showed a significantly positive association with the abundance of Th17 cells. Conversely, higher expression levels of LRPPRC were associated with a reduced abundance of various immune cells, particularly monocytes. CD patients were classified into two disulfidptosis-related subtypes. Cluster B patients exhibited lower immune infiltration and milder clinical presentation.
Conclusion: LRPPRC and NDUFA11 are identified as hub DRGs in CD, with potential roles in disulfidptosis and immune regulation. The disulfidptosis subtypes provide new insights into disease progression.
Keywords: Crohn’s disease; disulfidptosis; expression pattern; immune cell infiltration; machine learning.
© 2024 Fu et al.
Conflict of interest statement
The authors declare that they have no competing interests in this work.
Figures





Similar articles
-
Integrated bioinformatic analysis of immune infiltration and disulfidptosis related gene subgroups in type A aortic dissection.Sci Rep. 2025 Apr 21;15(1):13719. doi: 10.1038/s41598-025-98149-y. Sci Rep. 2025. PMID: 40258895 Free PMC article.
-
Characterization of moyamoya disease molecular subtypes through disulfidptosis‑related genes and immune landscape analysis.Exp Ther Med. 2025 Feb 14;29(4):74. doi: 10.3892/etm.2025.12824. eCollection 2025 Apr. Exp Ther Med. 2025. PMID: 40012919 Free PMC article.
-
Identification of disulfidptosis-related genes and analysis of immune infiltration characteristics in ischemic strokes.Math Biosci Eng. 2023 Oct 10;20(10):18939-18959. doi: 10.3934/mbe.2023838. Math Biosci Eng. 2023. PMID: 38052584
-
Identification of hub programmed cell death-related genes and immune infiltration in Crohn's disease using bioinformatics.Front Genet. 2024 Dec 18;15:1425062. doi: 10.3389/fgene.2024.1425062. eCollection 2024. Front Genet. 2024. PMID: 39744064 Free PMC article.
-
Disulfidptosis: a new target for central nervous system disease therapy.Front Neurosci. 2025 Mar 5;19:1514253. doi: 10.3389/fnins.2025.1514253. eCollection 2025. Front Neurosci. 2025. PMID: 40109666 Free PMC article. Review.
Cited by
-
Identification biomarkers and therapeutic targets of disulfidptosis-related in rheumatoid arthritis via bioinformatics, molecular dynamics simulation, and experimental validation.Sci Rep. 2025 Mar 13;15(1):8779. doi: 10.1038/s41598-025-93656-4. Sci Rep. 2025. PMID: 40082645 Free PMC article.
References
LinkOut - more resources
Full Text Sources

