Integrated transcriptomic and immunological profiling reveals new diagnostic and prognostic models for cutaneous melanoma
- PMID: 38863979
- PMCID: PMC11165152
- DOI: 10.3389/fphar.2024.1389550
Integrated transcriptomic and immunological profiling reveals new diagnostic and prognostic models for cutaneous melanoma
Abstract
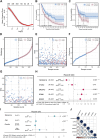
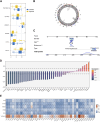
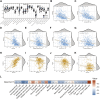
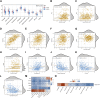
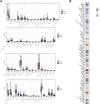
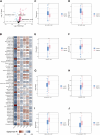
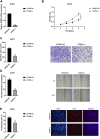
The mortality rate associated with cutaneous melanoma (SKCM) remains alarmingly high, highlighting the urgent need for a deeper understanding of its molecular underpinnings. In our study, we leveraged bulk transcriptome sequencing data from the SKCM cohort available in public databases such as TCGA and GEO. We utilized distinct datasets for training and validation purposes and also incorporated mutation and clinical data from TCGA, along with single-cell sequencing data from GEO. Through dimensionality reduction, we annotated cell subtypes within the single-cell data and analyzed the expression of tumor-related pathways across these subtypes. We identified differentially expressed genes (DEGs) in the training set, which were further refined using the Least Absolute Shrinkage and Selection Operator (LASSO) machine learning algorithm, employing tenfold cross-validation. This enabled the construction of a prognostic model, whose diagnostic efficacy we subsequently validated. We conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses on the DEGs, and performed immunological profiling on two risk groups to elucidate the relationship between model genes and the immune responses relevant to SKCM diagnosis, treatment, and prognosis. We also knocked down the GMR6 expression level in the melanoma cells and verified its effect on cancer through multiple experiments. The results indicate that the GMR6 gene plays a role in promoting the proliferation, invasion, and migration of cancer cells in human melanoma. Our findings offer novel insights and a theoretical framework that could enhance prognosis, treatment, and drug development strategies for SKCM, potentially leading to more precise therapeutic interventions.
Keywords: LASSO machine learning algorithm; SKCM; differential gene expression; immune infiltration analysis; melanoma.
Copyright © 2024 Li, Wu, Lin, Zhou and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Dhingra A., Fina M. E., Neinstein A., Ramsey D. J., Xu Y., Fishman G. A., et al. (2011). Autoantibodies in melanoma-associated retinopathy target TRPM1 cation channels of retinal ON bipolar cells. J. Neurosci. official J. Soc. Neurosci. 31 (11), 3962–3967. 10.1523/JNEUROSCI.6007-10.2011 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

