Coevolutionary analysis of Pseudomonas syringae-phage interactions to help with rational design of phage treatments
- PMID: 38864499
- PMCID: PMC11167607
- DOI: 10.1111/1751-7915.14489
Coevolutionary analysis of Pseudomonas syringae-phage interactions to help with rational design of phage treatments
Abstract
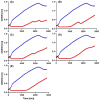
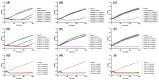
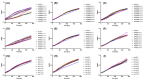
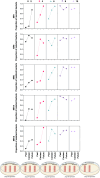
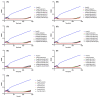
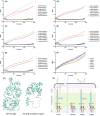
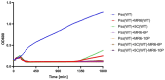
Treating plant bacterial diseases is notoriously difficult because of the lack of available antimicrobials. Pseudomonas syringae pathovar syringae (Pss) is a major pathogen of cherry (Prunus avium) causing bacterial canker of the stem, leaf and fruit, impacting productivity and leading to a loss of trees. In an attempt to find a treatment for this disease, naturally occurring bacteriophage (phage) that specifically target Pss is being investigated as a biocontrol strategy. However, before using them as a biocontrol treatment, it is important to both understand their efficacy in reducing the bacterial population and determine if the bacterial pathogens can evolve resistance to evade phage infection. To investigate this, killing curve assays of five MR phages targeting Pss showed that phage resistance rapidly emerges in vitro, even when using a cocktail of the five phages together. To gain insight to the changes occurring, Pss colonies were collected three times during a 66-h killing curve assay and separately, Pss and phage were also coevolved over 10 generations, enabling the measurement of genomic and fitness changes in bacterial populations. Pss evolved resistance to phages through modifications in lipopolysaccharide (LPS) synthesis pathways. Bacterial fitness (growth) and virulence were affected in only a few mutants. Deletion of LPS-associated genes suggested that LPS was the main target receptor for all five MR phages. Later generations of coevolved phages from the coevolution experiment were more potent at reducing the bacterial density and when used with wild-type phages could reduce the emergence of phage-resistant mutants. This study shows that understanding the genetic mechanisms of bacterial pathogen resistance to phages is important for helping to design a more effective approach to kill the bacteria while minimizing the opportunity for phage resistance to manifest.
© 2024 The Author(s). Microbial Biotechnology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Brüssow, H. (2005) Phage therapy: the Escherichia coli experience. Microbiology, 151(7), 2133–2140. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

