The biology and pathogenicity of Clostridium perfringens type F: a common human enteropathogen with a new(ish) name
- PMID: 38864615
- PMCID: PMC11426027
- DOI: 10.1128/mmbr.00140-23
The biology and pathogenicity of Clostridium perfringens type F: a common human enteropathogen with a new(ish) name
Abstract
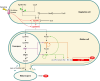
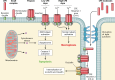
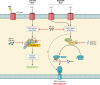
SUMMARYIn the 2018-revised Clostridium perfringens typing classification system, isolates carrying the enterotoxin (cpe) and alpha toxin genes but no other typing toxin genes are now designated as type F. Type F isolates cause food poisoning and nonfoodborne human gastrointestinal (GI) diseases, which most commonly involve type F isolates carrying, respectivefooly, a chromosomal or plasmid-borne cpe gene. Compared to spores of other C. perfringens isolates, spores of type F chromosomal cpe isolates often exhibit greater resistance to food environment stresses, likely facilitating their survival in improperly prepared or stored foods. Multiple factors contribute to this spore resistance phenotype, including the production of a variant small acid-soluble protein-4. The pathogenicity of type F isolates involves sporulation-dependent C. perfringens enterotoxin (CPE) production. C. perfringens sporulation is initiated by orphan histidine kinases and sporulation-associated sigma factors that drive cpe transcription. CPE-induced cytotoxicity starts when CPE binds to claudin receptors to form a small complex (which also includes nonreceptor claudins). Approximately six small complexes oligomerize on the host cell plasma membrane surface to form a prepore. CPE molecules in that prepore apparently extend β-hairpin loops to form a β-barrel pore, allowing a Ca2+ influx that activates calpain. With low-dose CPE treatment, caspase-3-dependent apoptosis develops, while high-CPE dose treatment induces necroptosis. Those effects cause histologic damage along with fluid and electrolyte losses from the colon and small intestine. Sialidases likely contribute to type F disease by enhancing CPE action and, for NanI-producing nonfoodborne human GI disease isolates, increasing intestinal growth and colonization.
Keywords: Clostridium perfringens; enterotoxin; germination; orphan histidine kinases; small acid-soluble proteins; sporulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops.Mol Microbiol. 1999 Sep;33(5):946-58. doi: 10.1046/j.1365-2958.1999.01534.x. Mol Microbiol. 1999. PMID: 10476029
-
NanH Is Produced by Sporulating Cultures of Clostridium perfringens Type F Food Poisoning Strains and Enhances the Cytotoxicity of C. perfringens Enterotoxin.mSphere. 2021 Apr 28;6(2):e00176-21. doi: 10.1128/mSphere.00176-21. mSphere. 2021. PMID: 33910991 Free PMC article.
-
Identification of an Important Orphan Histidine Kinase for the Initiation of Sporulation and Enterotoxin Production by Clostridium perfringens Type F Strain SM101.mBio. 2019 Jan 22;10(1):e02674-18. doi: 10.1128/mBio.02674-18. mBio. 2019. PMID: 30670619 Free PMC article.
-
Clostridium perfringens Enterotoxin: Action, Genetics, and Translational Applications.Toxins (Basel). 2016 Mar 16;8(3):73. doi: 10.3390/toxins8030073. Toxins (Basel). 2016. PMID: 26999202 Free PMC article. Review.
-
Enterotoxigenic Clostridium perfringens: detection and identification.Microbes Environ. 2012;27(4):343-9. doi: 10.1264/jsme2.me12002. Epub 2012 Apr 14. Microbes Environ. 2012. PMID: 22504431 Free PMC article. Review.
Cited by
-
Gut microbiota and risk of iron deficiency anemia: A two-sample Mendelian randomization study.Medicine (Baltimore). 2025 Feb 21;104(8):e41617. doi: 10.1097/MD.0000000000041617. Medicine (Baltimore). 2025. PMID: 39993092 Free PMC article.
-
Molecular Epidemiology of Type F Clostridium perfringens Among Diarrheal Patients and Virulence-Resistance Dynamics - 11 Provinces, China, 2024.China CDC Wkly. 2025 Jan 17;7(3):69-76. doi: 10.46234/ccdcw2025.013. China CDC Wkly. 2025. PMID: 39867821 Free PMC article.
-
Processing of Clostridium perfringens Enterotoxin by Intestinal Proteases.Toxins (Basel). 2025 Apr 1;17(4):170. doi: 10.3390/toxins17040170. Toxins (Basel). 2025. PMID: 40278668 Free PMC article.
-
Prevalence and toxin gene profiles of Clostridium perfringens in diarrheic dogs and cats in Korea: a retrospective analysis.J Vet Sci. 2025 Jul;26(4):e52. doi: 10.4142/jvs.24361. J Vet Sci. 2025. PMID: 40765232 Free PMC article.
-
Processing of Clostridium perfringens Enterotoxin by Intestinal Proteases.bioRxiv [Preprint]. 2025 Feb 11:2025.02.11.637699. doi: 10.1101/2025.02.11.637699. bioRxiv. 2025. Update in: Toxins (Basel). 2025 Apr 01;17(4):170. doi: 10.3390/toxins17040170. PMID: 39990433 Free PMC article. Updated. Preprint.
References
-
- McClane BA, Robertson SL, Li J. 2013. Clostridium perfringens, p 465–489. In Doyle MP, Buchanan RL (ed), Food microbiology: fundamentals and frontiers, 4th ed. ASM press, Washington D.C.
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI019844/AI/NIAID NIH HHS/United States
- R21 AI125796/AI/NIAID NIH HHS/United States
- R21 AI148911/AI/NIAID NIH HHS/United States
- R01AI019844-41/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- R21AI148911-02/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

