Comprehensive analysis of lncRNA-associated ceRNA network reveals novel potential prognostic regulatory axes in glioblastoma multiforme
- PMID: 38864705
- PMCID: PMC11167707
- DOI: 10.1111/jcmm.18392
Comprehensive analysis of lncRNA-associated ceRNA network reveals novel potential prognostic regulatory axes in glioblastoma multiforme
Abstract
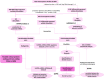
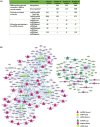
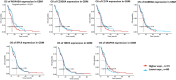
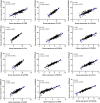
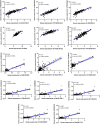
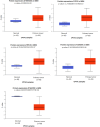
Deciphering the lncRNA-associated competitive endogenous RNA (ceRNA) network is essential in decoding glioblastoma multiforme (GBM) pathogenesis by regulating miRNA availability and controlling mRNA stability. This study aimed to explore novel biomarkers for GBM by constructing a lncRNA-miRNA-mRNA network. A ceRNA network in GBM was constructed using lncRNA, mRNA and miRNA expression profiles from the TCGA and GEO datasets. Seed nodes were identified by protein-protein interaction (PPI) network analysis of deregulated-mRNAs (DEmRNAs) in the ceRNA network. A lncRNA-miRNA-seed network was constructed by mapping the seed nodes into the preliminary ceRNA network. The impact of the seed nodes on the overall survival (OS) of patients was assessed by the GSCA database. Functional enrichment analysis of the deregulated-lncRNAs (DElncRNA) in the ceRNA network and genes interacting with OS-related genes in the PPI network were performed. Finally, the positive correlation between seed nodes and their associated lncRNAs and the expression level of these molecules in GBM tissue compared with normal samples was validated using the GEPIA database. Our analyzes revealed that three novel regulatory axes AL161785.1/miR-139-5p/MS4A6A, LINC02611/miR-139-5p/MS4A6A and PCED1B-AS1/miR-433-3p/MS4A6A may play essential roles in GBM pathogenesis. MS4A6A is upregulated in GBM and closely associated with shorter survival time of patients. We also identified that MS4A6A expression positively correlates with genes related to tumour-associated macrophages, which induce macrophage infiltration and immune suppression. The functional enrichment analysis demonstrated that DElncRNAs are mainly involved in neuroactive ligand-receptor interaction, calcium/MAPK signalling pathway, ribosome, GABAergic/Serotonergic/Glutamatergic synapse and immune system process. In addition, genes related to MS4A6A contribute to immune and inflammatory-related biological processes. Our findings provide novel insights to understand the ceRNA regulation in GBM and identify novel prognostic biomarkers or therapeutic targets.
Keywords: AL161785.1; LINC02611; MS4A6A; PCED1B‐AS1; glioblastoma multiforme; miR‐139‐5p; miR‐433‐3p.
© 2024 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Chen Q, Cai J, Wang Q, et al. Long noncoding RNA NEAT1, regulated by the EGFR pathway, contributes to glioblastoma progression through the WNT/β‐catenin pathway by scaffolding EZH2Oncogenic function of NEAT1 in glioblastoma. Clin Cancer Res. 2018;24:684‐695. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

