Molecular basis for different substrate-binding sites and chaperone functions of the BRICHOS domain
- PMID: 38864729
- PMCID: PMC11168071
- DOI: 10.1002/pro.5063
Molecular basis for different substrate-binding sites and chaperone functions of the BRICHOS domain
Abstract
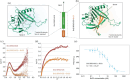
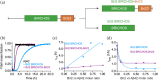
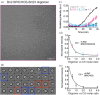
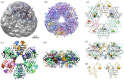
Proteins can misfold into fibrillar or amorphous aggregates and molecular chaperones act as crucial guardians against these undesirable processes. The BRICHOS chaperone domain, found in several otherwise unrelated proproteins that contain amyloidogenic regions, effectively inhibits amyloid formation and toxicity but can in some cases also prevent non-fibrillar, amorphous protein aggregation. Here, we elucidate the molecular basis behind the multifaceted chaperone activities of the BRICHOS domain from the Bri2 proprotein. High-confidence AlphaFold2 and RoseTTAFold predictions suggest that the intramolecular amyloidogenic region (Bri23) is part of the hydrophobic core of the proprotein, where it occupies the proposed amyloid binding site, explaining the markedly reduced ability of the proprotein to prevent an exogenous amyloidogenic peptide from aggregating. However, the BRICHOS-Bri23 complex maintains its ability to form large polydisperse oligomers that prevent amorphous protein aggregation. A cryo-EM-derived model of the Bri2 BRICHOS oligomer is compatible with surface-exposed hydrophobic motifs that get exposed and come together during oligomerization, explaining its effects against amorphous aggregation. These findings provide a molecular basis for the BRICHOS chaperone domain function, where distinct surfaces are employed against different forms of protein aggregation.
Keywords: BRICHOS; fibrillar and amorphous aggregation; molecular chaperone; oligomer model.
© 2024 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Short hydrophobic loop motifs in BRICHOS domains determine chaperone activity against amorphous protein aggregation but not against amyloid formation.Commun Biol. 2023 May 8;6(1):497. doi: 10.1038/s42003-023-04883-2. Commun Biol. 2023. PMID: 37156997 Free PMC article.
-
Recombinant Bri3 BRICHOS domain is a molecular chaperone with effect against amyloid formation and non-fibrillar protein aggregation.Sci Rep. 2020 Jun 17;10(1):9817. doi: 10.1038/s41598-020-66718-y. Sci Rep. 2020. PMID: 32555390 Free PMC article.
-
Specific inhibition of α-synuclein oligomer generation and toxicity by the chaperone domain Bri2 BRICHOS.Protein Sci. 2024 Aug;33(8):e5091. doi: 10.1002/pro.5091. Protein Sci. 2024. PMID: 38980078 Free PMC article.
-
A new kid in the folding funnel: Molecular chaperone activities of the BRICHOS domain.Protein Sci. 2023 Jun;32(6):e4645. doi: 10.1002/pro.4645. Protein Sci. 2023. PMID: 37096906 Free PMC article. Review.
-
BRICHOS domain associated with lung fibrosis, dementia and cancer--a chaperone that prevents amyloid fibril formation?FEBS J. 2011 Oct;278(20):3893-904. doi: 10.1111/j.1742-4658.2011.08209.x. Epub 2011 Jul 5. FEBS J. 2011. PMID: 21668643 Review.
Cited by
-
Determinants for Substoichiometric Inhibition of IAPP and Aβ Amyloid Aggregations by Bri2 BRICHOS.ACS Chem Neurosci. 2025 Mar 19;16(6):1150-1160. doi: 10.1021/acschemneuro.4c00839. Epub 2025 Mar 4. ACS Chem Neurosci. 2025. PMID: 40035576
-
Chaperone-Mediated Regulation of Tau Phase Separation, Fibrillation, and Toxicity.J Am Chem Soc. 2025 Jul 9;147(27):23504-23518. doi: 10.1021/jacs.5c01369. Epub 2025 Jun 29. J Am Chem Soc. 2025. PMID: 40583215 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- Alzheimer's Association Research Grant (U.S.)
- Vetenskapsrådet
- Olle Engkvists Stiftelse
- Petrus and Augusta Hedlunds Stiftelse
- Åke Wibergs stiftelse
- the Swedish Alzheimer Foundation
- Åhlén Stiftelsens
- Karolinska Institutet Research Foundation Grant
- Stiftelsen för Gamla Tjänarinnor
- Stiftelsen Sigurd och Elsa Goljes Minne
- Loo and Hans Osterman Foundation for Medical Research
- Geriatric Diseases Foundation at Karolinska Institutet
- Gun and Bertil Stohne's Foundation
- Magnus Bergvall Foundation
- FORMAS
- China Scholarship Council
- China Association for Science and Technology
LinkOut - more resources
Full Text Sources
Miscellaneous

