DBF4, not DRF1, is the crucial regulator of CDC7 kinase at replication forks
- PMID: 38865090
- PMCID: PMC11169917
- DOI: 10.1083/jcb.202402144
DBF4, not DRF1, is the crucial regulator of CDC7 kinase at replication forks
Abstract
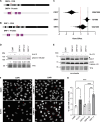
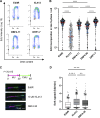
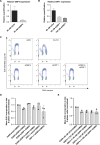
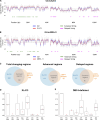
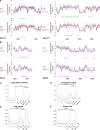
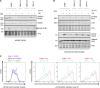
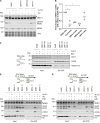
CDC7 kinase is crucial for DNA replication initiation and is involved in fork processing and replication stress response. Human CDC7 requires the binding of either DBF4 or DRF1 for its activity. However, it is unclear whether the two regulatory subunits target CDC7 to a specific set of substrates, thus having different biological functions, or if they act redundantly. Using genome editing technology, we generated isogenic cell lines deficient in either DBF4 or DRF1: these cells are viable but present signs of genomic instability, indicating that both can independently support CDC7 for bulk DNA replication. Nonetheless, DBF4-deficient cells show altered replication efficiency, partial deficiency in MCM helicase phosphorylation, and alterations in the replication timing of discrete genomic regions. Notably, we find that CDC7 function at replication forks is entirely dependent on DBF4 and not on DRF1. Thus, DBF4 is the primary regulator of CDC7 activity, mediating most of its functions in unperturbed DNA replication and upon replication interference.
© 2024 Göder et al.
Conflict of interest statement
Disclosures: A. Göder reported personal fees from AstraZeneca outside the submitted work; and “While unrelated to the submitted work, A. Göder would like to disclose that she is currently employed by AstraZeneca and has stock ownership and/or stock options or interests in the company.” C. Santocanale reported personal fees from Turbine LTD outside the submitted work; in addition, C. Santocanale had a patent to EP4275686A1 pending. No other disclosures were reported.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

