Meiosis-specific functions of kinetochore protein SPC105R required for chromosome segregation in Drosophila oocytes
- PMID: 38865189
- PMCID: PMC11321039
- DOI: 10.1091/mbc.E24-02-0067
Meiosis-specific functions of kinetochore protein SPC105R required for chromosome segregation in Drosophila oocytes
Abstract
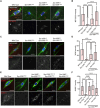
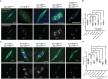
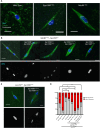
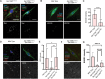
The reductional division of meiosis I requires the separation of chromosome pairs towards opposite poles. We have previously implicated the outer kinetochore protein SPC105R/KNL1 in driving meiosis I chromosome segregation through lateral attachments to microtubules and coorientation of sister centromeres. To identify the domains of SPC105R that are critical for meiotic chromosome segregation, an RNAi-resistant gene expression system was developed. We found that the SPC105R C-terminal domain (aa 1284-1960) is necessary and sufficient for recruiting NDC80 to the kinetochore and building the outer kinetochore. Furthermore, the C-terminal domain recruits BUBR1, which in turn recruits the cohesion protection proteins MEI-S332 and PP2A. Of the remaining 1283 amino acids, we found the first 473 are most important for meiosis. The first 123 amino acids of the N-terminal half of SPC105R contain the conserved SLRK and RISF motifs that are targets of PP1 and Aurora B kinase and are most important for regulating the stability of microtubule attachments and maintaining metaphase I arrest. The region between amino acids 124 and 473 are required for lateral microtubule attachments and biorientation of homologues, which are critical for accurate chromosome segregation in meiosis I.
Figures




Update of
-
Meiosis-specific functions of kinetochore protein SPC105R required for chromosome segregation in Drosophila oocytes.bioRxiv [Preprint]. 2024 Mar 14:2024.03.14.585003. doi: 10.1101/2024.03.14.585003. bioRxiv. 2024. Update in: Mol Biol Cell. 2024 Aug 1;35(8):ar105. doi: 10.1091/mbc.E24-02-0067. PMID: 38559067 Free PMC article. Updated. Preprint.
Similar articles
-
Meiosis-specific functions of kinetochore protein SPC105R required for chromosome segregation in Drosophila oocytes.bioRxiv [Preprint]. 2024 Mar 14:2024.03.14.585003. doi: 10.1101/2024.03.14.585003. bioRxiv. 2024. Update in: Mol Biol Cell. 2024 Aug 1;35(8):ar105. doi: 10.1091/mbc.E24-02-0067. PMID: 38559067 Free PMC article. Updated. Preprint.
-
Sister centromere fusion during meiosis I depends on maintaining cohesins and destabilizing microtubule attachments.PLoS Genet. 2019 May 31;15(5):e1008072. doi: 10.1371/journal.pgen.1008072. eCollection 2019 May. PLoS Genet. 2019. PMID: 31150390 Free PMC article.
-
Regulation of centromere localization of the Drosophila Shugoshin MEI-S332 and sister-chromatid cohesion in meiosis.G3 (Bethesda). 2014 Jul 31;4(10):1849-58. doi: 10.1534/g3.114.012823. G3 (Bethesda). 2014. PMID: 25081981 Free PMC article.
-
Mechanisms of kinesin-7 CENP-E in kinetochore-microtubule capture and chromosome alignment during cell division.Biol Cell. 2019 Jun;111(6):143-160. doi: 10.1111/boc.201800082. Epub 2019 Feb 26. Biol Cell. 2019. PMID: 30784092 Review.
-
The Molecular Mechanism of Aurora-B Regulating Kinetochore-Microtubule Attachment in Mitosis and Oocyte Meiosis.Cytogenet Genome Res. 2024;164(2):69-77. doi: 10.1159/000540588. Epub 2024 Jul 27. Cytogenet Genome Res. 2024. PMID: 39068909 Review.
Cited by
-
The functional organisation of the centromere and kinetochore during meiosis.Curr Opin Cell Biol. 2025 Jun;94:102486. doi: 10.1016/j.ceb.2025.102486. Epub 2025 Feb 26. Curr Opin Cell Biol. 2025. PMID: 40015116 Free PMC article. Review.
-
Distinct checkpoint and homolog biorientation pathways regulate meiosis I in Drosophila oocytes.PLoS Genet. 2025 Jan 29;21(1):e1011400. doi: 10.1371/journal.pgen.1011400. eCollection 2025 Jan. PLoS Genet. 2025. PMID: 39879252 Free PMC article.
-
Distinct checkpoint and homolog biorientation pathways regulate meiosis I in Drosophila oocytes.bioRxiv [Preprint]. 2024 Aug 21:2024.08.21.608908. doi: 10.1101/2024.08.21.608908. bioRxiv. 2024. Update in: PLoS Genet. 2025 Jan 29;21(1):e1011400. doi: 10.1371/journal.pgen.1011400. PMID: 39229242 Free PMC article. Updated. Preprint.
References
-
- Bischof J, Bjorklund M, Furger E, Schertel C, Taipale J, Basler K (2013). A versatile platform for creating a comprehensive UAS-ORFeome library in Drosophila. Development 140, 2434–2442. - PubMed
-
- Bonner AM, Hughes SE, Chisholm JA, Smith SK, Slaughter BD, Unruh JR, Collins KA, Friederichs JM, Florens L, Swanson SK, et al. (2013). Binding of Drosophila Polo kinase to its regulator Matrimony is noncanonical and involves two separate functional domains. Proc Natl Acad Sci USA 110, E1222–E1231. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

