CGG repeats in the human FMR1 gene regulate mRNA localization and cellular stress in developing neurons
- PMID: 38865241
- PMCID: PMC11240841
- DOI: 10.1016/j.celrep.2024.114330
CGG repeats in the human FMR1 gene regulate mRNA localization and cellular stress in developing neurons
Abstract
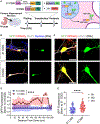
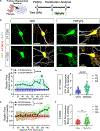
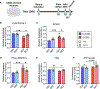
The human genome has many short tandem repeats, yet the normal functions of these repeats are unclear. The 5' untranslated region (UTR) of the fragile X messenger ribonucleoprotein 1 (FMR1) gene contains polymorphic CGG repeats, the length of which has differing effects on FMR1 expression and human health, including the neurodevelopmental disorder fragile X syndrome. We deleted the CGG repeats in the FMR1 gene (0CGG) in human stem cells and examined the effects on differentiated neurons. 0CGG neurons have altered subcellular localization of FMR1 mRNA and protein, and differential expression of cellular stress proteins compared with neurons with normal repeats (31CGG). In addition, 0CGG neurons have altered responses to glucocorticoid receptor (GR) activation, including FMR1 mRNA localization, GR chaperone HSP90α expression, GR localization, and cellular stress protein levels. Therefore, the CGG repeats in the FMR1 gene are important for the homeostatic responses of neurons to stress signals.
Keywords: 5′ UTR; CGG repeats; CP: Molecular biology; CP: Neuroscience; FMR1; FMRP; HSP90; RNA localization; dexamethasone; fragile X syndrome; glucocorticoid receptor; human-specific; neuron; pluripotent stem cells; single molecule FISH; stress; trinucleotide repeats.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
FMR1 CGG repeat lengths mediate different regulation of reporter gene expression in comparative transient and locus specific integration assays.Gene. 2011 Oct 15;486(1-2):15-22. doi: 10.1016/j.gene.2011.06.034. Epub 2011 Jul 13. Gene. 2011. PMID: 21767618
-
Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome.PLoS One. 2011;6(10):e26203. doi: 10.1371/journal.pone.0026203. Epub 2011 Oct 12. PLoS One. 2011. PMID: 22022567 Free PMC article.
-
CGG-repeat dynamics and FMR1 gene silencing in fragile X syndrome stem cells and stem cell-derived neurons.Mol Autism. 2016 Oct 6;7:42. doi: 10.1186/s13229-016-0105-9. eCollection 2016. Mol Autism. 2016. PMID: 27713816 Free PMC article.
-
Spontaneous rescue of a FMR1 repeat expansion and review of deletions in the FMR1 non-coding region.Eur J Med Genet. 2021 Aug;64(8):104244. doi: 10.1016/j.ejmg.2021.104244. Epub 2021 May 20. Eur J Med Genet. 2021. PMID: 34022415 Review.
-
[Chromatin changes caused by CGG repeat expansion in fmr1 gene].Mol Biol (Mosk). 2015 Mar-Apr;49(2):205-11. Mol Biol (Mosk). 2015. PMID: 26065250 Review. Russian.
Cited by
-
FMRP gains mitochondrial fission control.Nat Cell Biol. 2024 Dec;26(12):2014-2015. doi: 10.1038/s41556-024-01567-9. Nat Cell Biol. 2024. PMID: 39548328 No abstract available.
-
FMR1: A Neurodevelopmental Factor Regulating Cell Metabolism in the Tumor Microenvironment.Biomolecules. 2025 May 28;15(6):779. doi: 10.3390/biom15060779. Biomolecules. 2025. PMID: 40563420 Free PMC article. Review.
-
Trinucleotide repeat expansion and RNA dysregulation in fragile X syndrome: emerging therapeutic approaches.RNA. 2025 Feb 19;31(3):307-313. doi: 10.1261/rna.080270.124. RNA. 2025. PMID: 39725461 Free PMC article. Review.
-
Beyond the Synapse: FMR1 and FMRP Molecular Mechanisms in the Nucleus.Int J Mol Sci. 2024 Dec 30;26(1):214. doi: 10.3390/ijms26010214. Int J Mol Sci. 2024. PMID: 39796070 Free PMC article. Review.
References
-
- Rafehi H, Read J, Szmulewicz DJ, Davies KC, Snell P, Fearnley LG, Scott L, Thomsen M, Gillies G, Pope K, et al. (2023). An intronic GAA repeat expansion in FGF14 causes the autosomal-dominant adult-onset ataxia SCA27B/ATX-FGF14. Am. J. Hum. Genet. 110, 1018. 10.1016/j.ajhg.2023.05.005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

