RNF212B E3 ligase is essential for crossover designation and maturation during male and female meiosis in the mouse
- PMID: 38865271
- PMCID: PMC11194559
- DOI: 10.1073/pnas.2320995121
RNF212B E3 ligase is essential for crossover designation and maturation during male and female meiosis in the mouse
Erratum in
-
Correction for Condezo et al., RNF212B E3 ligase is essential for crossover designation and maturation during male and female meiosis in the mouse.Proc Natl Acad Sci U S A. 2024 Oct 8;121(41):e2418611121. doi: 10.1073/pnas.2418611121. Epub 2024 Sep 30. Proc Natl Acad Sci U S A. 2024. PMID: 39348550 Free PMC article. No abstract available.
Abstract
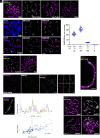
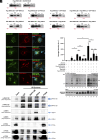
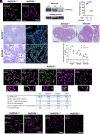
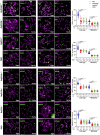
Meiosis, a reductional cell division, relies on precise initiation, maturation, and resolution of crossovers (COs) during prophase I to ensure the accurate segregation of homologous chromosomes during metaphase I. This process is regulated by the interplay of RING-E3 ligases such as RNF212 and HEI10 in mammals. In this study, we functionally characterized a recently identified RING-E3 ligase, RNF212B. RNF212B colocalizes and interacts with RNF212, forming foci along chromosomes from zygonema onward in a synapsis-dependent and DSB-independent manner. These consolidate into larger foci at maturing COs, colocalizing with HEI10, CNTD1, and MLH1 by late pachynema. Genetically, RNF212B foci formation depends on Rnf212 but not on Msh4, Hei10, and Cntd1, while the unloading of RNF212B at the end of pachynema is dependent on Hei10 and Cntd1. Mice lacking RNF212B, or expressing an inactive RNF212B protein, exhibit modest synapsis defects, a reduction in the localization of pro-CO factors (MSH4, TEX11, RPA, MZIP2) and absence of late CO-intermediates (MLH1). This loss of most COs by diakinesis results in mostly univalent chromosomes. Double mutants for Rnf212b and Rnf212 exhibit an identical phenotype to that of Rnf212b single mutants, while double heterozygous demonstrate a dosage-dependent reduction in CO number, indicating a functional interplay between paralogs. SUMOylome analysis of testes from Rnf212b mutants and pull-down analysis of Sumo- and Ubiquitin-tagged HeLa cells, suggest that RNF212B is an E3-ligase with Ubiquitin activity, serving as a crucial factor for CO maturation. Thus, RNF212 and RNF212B play vital, yet overlapping roles, in ensuring CO homeostasis through their distinct E3 ligase activities.
Keywords: crossing over; fertility; meiosis; mouse; recombination.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

