Histone ADP-ribosylation promotes resistance to PARP inhibitors by facilitating PARP1 release from DNA lesions
- PMID: 38865276
- PMCID: PMC11194589
- DOI: 10.1073/pnas.2322689121
Histone ADP-ribosylation promotes resistance to PARP inhibitors by facilitating PARP1 release from DNA lesions
Abstract
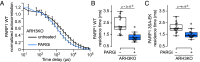
Poly(ADP-ribose) polymerase 1 (PARP1) has emerged as a central target for cancer therapies due to the ability of PARP inhibitors to specifically kill tumors deficient for DNA repair by homologous recombination. Upon DNA damage, PARP1 quickly binds to DNA breaks and triggers ADP-ribosylation signaling. ADP-ribosylation is important for the recruitment of various factors to sites of damage, as well as for the timely dissociation of PARP1 from DNA breaks. Indeed, PARP1 becomes trapped at DNA breaks in the presence of PARP inhibitors, a mechanism underlying the cytotoxitiy of these inhibitors. Therefore, any cellular process influencing trapping is thought to impact PARP inhibitor efficiency, potentially leading to acquired resistance in patients treated with these drugs. There are numerous ADP-ribosylation targets after DNA damage, including PARP1 itself as well as histones. While recent findings reported that the automodification of PARP1 promotes its release from the DNA lesions, the potential impact of other ADP-ribosylated proteins on this process remains unknown. Here, we demonstrate that histone ADP-ribosylation is also crucial for the timely dissipation of PARP1 from the lesions, thus contributing to cellular resistance to PARP inhibitors. Considering the crosstalk between ADP-ribosylation and other histone marks, our findings open interesting perspectives for the development of more efficient PARP inhibitor-driven cancer therapies.
Keywords: ADP-ribosylation; DNA repair; PARP1; chromatin; fluorescence microscopy.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
MeSH terms
Substances
Grants and funding
- CD53/Ligue Contre le Cancer (French League Against Cancer)
- ANR-22-CE12-0039/Agence Nationale de la Recherche (ANR)
- ARCDOC42022010004560/Fondation ARC pour la Recherche sur le Cancer (ARC)
- 223107/WT_/Wellcome Trust/United Kingdom
- BB/R007195/1/UKRI | Biotechnology and Biological Sciences Research Council (BBSRC)
- PLBIO-2019/Institut National Du Cancer (INCa)
- C35050/A22284/Cancer Research United Kingdom
- ARCPJA2022060005190/Fondation ARC pour la Recherche sur le Cancer (ARC)
- 813369/Ovarian Cancer Research Alliance (OCRA)
- 210634/WT_/Wellcome Trust/United Kingdom
- 22284/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- PDF20181208405/Fondation ARC pour la Recherche sur le Cancer (ARC)
LinkOut - more resources
Full Text Sources
Miscellaneous

