Green/red light-sensing mechanism in the chromatic acclimation photosensor
- PMID: 38865454
- PMCID: PMC11168458
- DOI: 10.1126/sciadv.adn8386
Green/red light-sensing mechanism in the chromatic acclimation photosensor
Abstract
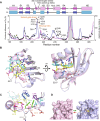
Certain cyanobacteria alter their photosynthetic light absorption between green and red, a phenomenon called complementary chromatic acclimation. The acclimation is regulated by a cyanobacteriochrome-class photosensor that reversibly photoconverts between green-absorbing (Pg) and red-absorbing (Pr) states. Here, we elucidated the structural basis of the green/red photocycle. In the Pg state, the bilin chromophore adopted the extended C15-Z,anti structure within a hydrophobic pocket. Upon photoconversion to the Pr state, the bilin is isomerized to the cyclic C15-E,syn structure, forming a water channel in the pocket. The solvation/desolvation of the bilin causes changes in the protonation state and the stability of π-conjugation at the B ring, leading to a large absorption shift. These results advance our understanding of the enormous spectral diversity of the phytochrome superfamily.
Figures


References
-
- Fushimi K., Narikawa R., Cyanobacteriochromes: photoreceptors covering the entire UV-to-visible spectrum. Curr. Opin. Struct. Biol. 57, 39–46 (2019). - PubMed
-
- Anders K., Essen L. O., The family of phytochrome-like photoreceptors: diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 35, 7–16 (2015). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

