TERRA transcripts localize at long telomeres to regulate telomerase access to chromosome ends
- PMID: 38865460
- PMCID: PMC11168465
- DOI: 10.1126/sciadv.adk4387
TERRA transcripts localize at long telomeres to regulate telomerase access to chromosome ends
Abstract
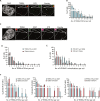
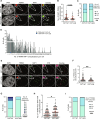
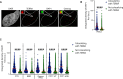
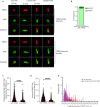
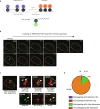
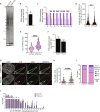
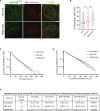
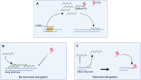
The function of TERRA in the regulation of telomerase in human cells is still debated. While TERRA interacts with telomerase, how it regulates telomerase function remains unknown. Here, we show that TERRA colocalizes with the telomerase RNA subunit hTR in the nucleoplasm and at telomeres during different phases of the cell cycle. We report that TERRA transcripts relocate away from chromosome ends during telomere lengthening, leading to a reduced number of telomeric TERRA-hTR molecules and consequent increase in "TERRA-free" telomerase molecules at telomeres. Using live-cell imaging and super-resolution microscopy, we show that upon transcription, TERRA relocates from its telomere of origin to long chromosome ends. Furthermore, TERRA depletion by antisense oligonucleotides promoted hTR localization to telomeres, leading to increased residence time and extended half-life of hTR molecules at telomeres. Overall, our findings indicate that telomeric TERRA transcripts inhibit telomere elongation by telomerase acting in trans, impairing telomerase access to telomeres that are different from their chromosome end of origin.
Figures

Similar articles
-
Telomere Length and Telomerase Activity as Biomarkers in the Diagnostics and Prognostics of Pathological Conditions.Biochemistry (Mosc). 2025 Jun;90(6):700-724. doi: 10.1134/S0006297925600814. Biochemistry (Mosc). 2025. PMID: 40609992 Review.
-
Telomere-specific regulation of TERRA and its impact on telomere stability.Semin Cell Dev Biol. 2024 Apr;157:3-23. doi: 10.1016/j.semcdb.2023.11.001. Epub 2023 Dec 11. Semin Cell Dev Biol. 2024. PMID: 38088000 Review.
-
The canonical RPA complex interacts with Est3 to regulate yeast telomerase activity.Proc Natl Acad Sci U S A. 2025 Feb 18;122(7):e2419309122. doi: 10.1073/pnas.2419309122. Epub 2025 Feb 6. Proc Natl Acad Sci U S A. 2025. PMID: 39913192 Free PMC article.
-
Comparison of Telomere Structure in Eukaryotes.Arch Razi Inst. 2024 Dec 31;79(6):1365-1374. doi: 10.32592/ARI.2024.79.6.1365. eCollection 2024 Dec. Arch Razi Inst. 2024. PMID: 40606259 Free PMC article. Review.
-
TPP1 OB-fold domain protein suppresses cell proliferation and induces cell apoptosis by inhibiting telomerase recruitment to telomeres in human lung cancer cells.J Cancer Res Clin Oncol. 2019 Jun;145(6):1509-1519. doi: 10.1007/s00432-019-02921-3. Epub 2019 Apr 23. J Cancer Res Clin Oncol. 2019. PMID: 31016380 Free PMC article.
Cited by
-
Telomeres: an organized string linking plants and mammals.Biol Direct. 2024 Nov 20;19(1):119. doi: 10.1186/s13062-024-00558-y. Biol Direct. 2024. PMID: 39568075 Free PMC article. Review.
-
RNA binding proteins (RBPs) on genetic stability and diseases.Glob Med Genet. 2024 Nov 30;12(1):100032. doi: 10.1016/j.gmg.2024.100032. eCollection 2025 Mar. Glob Med Genet. 2024. PMID: 39925443 Free PMC article. Review.
References
-
- Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J., Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 (2007). - PubMed
-
- Schoeftner S., Blasco M. A., Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236 (2008). - PubMed
-
- Luke B., Panza A., Redon S., Iglesias N., Li Z., Lingner J., The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell 32, 465–477 (2008). - PubMed
-
- Diman A., Decottignies A., Genomic origin and nuclear localization of TERRA telomeric repeat-containing RNA: From darkness to dawn. FEBS J. 285, 1389–1398 (2018). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

