Genetic diversity in the partial sequence of the HIV-1 gag gene among people living with multidrug-resistant HIV-1 infection
- PMID: 38865573
- PMCID: PMC11164046
- DOI: 10.1590/S1678-9946202466035
Genetic diversity in the partial sequence of the HIV-1 gag gene among people living with multidrug-resistant HIV-1 infection
Abstract
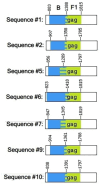
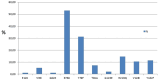
The group-specific antigen (gag) plays a crucial role in the assembly, release, and maturation of HIV. This study aimed to analyze the partial sequence of the HIV gag gene to classify HIV subtypes, identify recombination sites, and detect protease inhibitor (PI) resistance-associated mutations (RAMs). The cohort included 100 people living with HIV (PLH) who had experienced antiretroviral treatment failure with reverse transcriptase/protease inhibitors. Proviral HIV-DNA was successfully sequenced in 96 out of 100 samples for gag regions, specifically matrix (p17) and capsid (p24). Moreover, from these 96 sequences, 82 (85.42%) were classified as subtype B, six (6.25%) as subtype F1, one (1.04%) as subtype C, and seven (7.29%) exhibited a mosaic pattern between subtypes B and F1 (B/F1), with breakpoints at p24 protein. Insertions and deletions of amino acid at p17 were observed in 51 samples (53.13%). The prevalence of PI RAM in the partial gag gene was observed in 78 out of 96 PLH (81.25%). Among these cases, the most common mutations were R76K (53.13%), Y79F (31.25%), and H219Q (14.58%) at non-cleavage sites, as well as V128I (10.42%) and Y132F (11.46%) at cleavage sites. While B/F1 recombination was identified in the p24, the p17 coding region showed higher diversity, where insertions, deletions, and PI RAM, were observed at high prevalence. In PLH with virological failure, the analysis of the partial gag gene could contribute to more accurate predictions in genotypic resistance to PIs. This can aid guide more effective HIV treatment strategies.
Conflict of interest statement
The authors have declared that they do not have any competing interests.
Figures
Similar articles
-
In Vivo Emergence of a Novel Protease Inhibitor Resistance Signature in HIV-1 Matrix.mBio. 2020 Nov 3;11(6):e02036-20. doi: 10.1128/mBio.02036-20. mBio. 2020. PMID: 33144375 Free PMC article.
-
Positive selection pressure introduces secondary mutations at Gag cleavage sites in human immunodeficiency virus type 1 harboring major protease resistance mutations.J Virol. 2009 Sep;83(17):8916-24. doi: 10.1128/JVI.00003-09. Epub 2009 Jun 10. J Virol. 2009. PMID: 19515784 Free PMC article.
-
Gag P2/NC and pol genetic diversity, polymorphism, and drug resistance mutations in HIV-1 CRF02_AG- and non-CRF02_AG-infected patients in Yaoundé, Cameroon.Sci Rep. 2017 Oct 26;7(1):14136. doi: 10.1038/s41598-017-14095-4. Sci Rep. 2017. PMID: 29074854 Free PMC article.
-
Gag mutations can impact virological response to dual-boosted protease inhibitor combinations in antiretroviral-naïve HIV-infected patients.Antimicrob Agents Chemother. 2010 Jul;54(7):2910-9. doi: 10.1128/AAC.00194-10. Epub 2010 May 3. Antimicrob Agents Chemother. 2010. PMID: 20439606 Free PMC article. Clinical Trial.
-
Study of the impact of HIV genotypic drug resistance testing on therapy efficacy.Verh K Acad Geneeskd Belg. 2001;63(5):447-73. Verh K Acad Geneeskd Belg. 2001. PMID: 11813503 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

