Conformational Dynamics of the Nucleosomal Histone H2B Tails Revealed by Molecular Dynamics Simulations
- PMID: 38865599
- PMCID: PMC11200259
- DOI: 10.1021/acs.jcim.4c00059
Conformational Dynamics of the Nucleosomal Histone H2B Tails Revealed by Molecular Dynamics Simulations
Abstract
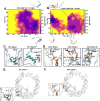
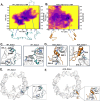
Epigenetic modifications of histone N-terminal tails play a critical role in regulating the chromatin structure and biological processes such as transcription and DNA repair. One of the key post-translational modifications (PTMs) is the acetylation of lysine residues on histone tails. Epigenetic modifications are ubiquitous in the development of diseases, such as cancer and neurological disorders. Histone H2B tails are critical regulators of nucleosome dynamics, biological processes, and certain diseases. Here, we report all-atomistic molecular dynamics (MD) simulations of the nucleosome to demonstrate that acetylation of the histone tails changes their conformational space and interaction with DNA. We perform simulations of H2B tails, critical regulators of gene regulation, in both the lysine-acetylated (ACK) and unacetylated wild type (WT) states. To explore the effects of salt concentration, we use two different NaCl concentrations to perform simulations at microsecond time scales. Salt can modulate the effects of electrostatic interactions between the DNA phosphate backbone and histone tails. Upon acetylation, H2B tails shift their secondary structure helical propensity. The number of contacts between the DNA and the H2B tail decreases. We characterize the conformational dynamics of the H2B tails by principal component analysis (PCA). The ACK tails become more compact at increased salt concentrations, but conformations from the WT tails display the most contacts with DNA at both salt concentrations. Mainly, H2B acetylation may increase the DNA accessibility for regulatory proteins to bind, which can aid in gene regulation and NCP stability.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

