PSMD11 loss-of-function variants correlate with a neurobehavioral phenotype, obesity, and increased interferon response
- PMID: 38866022
- PMCID: PMC11267520
- DOI: 10.1016/j.ajhg.2024.05.016
PSMD11 loss-of-function variants correlate with a neurobehavioral phenotype, obesity, and increased interferon response
Abstract
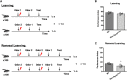
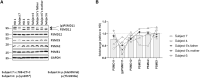
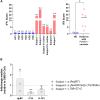
Primary proteasomopathies have recently emerged as a new class of rare early-onset neurodevelopmental disorders (NDDs) caused by pathogenic variants in the PSMB1, PSMC1, PSMC3, or PSMD12 proteasome genes. Proteasomes are large multi-subunit protein complexes that maintain cellular protein homeostasis by clearing ubiquitin-tagged damaged, misfolded, or unnecessary proteins. In this study, we have identified PSMD11 as an additional proteasome gene in which pathogenic variation is associated with an NDD-causing proteasomopathy. PSMD11 loss-of-function variants caused early-onset syndromic intellectual disability and neurodevelopmental delay with recurrent obesity in 10 unrelated children. Our findings demonstrate that the cognitive impairment observed in these individuals could be recapitulated in Drosophila melanogaster with depletion of the PMSD11 ortholog Rpn6, which compromised reversal learning. Our investigations in subject samples further revealed that PSMD11 loss of function resulted in impaired 26S proteasome assembly and the acquisition of a persistent type I interferon (IFN) gene signature, mediated by the integrated stress response (ISR) protein kinase R (PKR). In summary, these data identify PSMD11 as an additional member of the growing family of genes associated with neurodevelopmental proteasomopathies and provide insights into proteasomal biology in human health.
Copyright © 2024 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.C. and K.M. are employees of GeneDx, LLC. J.R.L. has stock in 23andMe and is a paid consultant for Genome International.
Figures





References
-
- Schmid H.P., Akhayat O., Martins De Sa C., Puvion F., Koehler K., Scherrer K. The prosome: an ubiquitous morphologically distinct RNP particle associated with repressed mRNPs and containing specific ScRNA and a characteristic set of proteins. EMBO J. 1984;3:29–34. doi: 10.1002/j.1460-2075.1984.tb01757.x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

