A multimodal generative AI copilot for human pathology
- PMID: 38866050
- PMCID: PMC11464372
- DOI: 10.1038/s41586-024-07618-3
A multimodal generative AI copilot for human pathology
Abstract
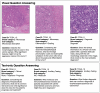
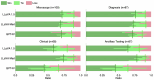
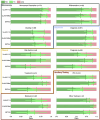
Computational pathology1,2 has witnessed considerable progress in the development of both task-specific predictive models and task-agnostic self-supervised vision encoders3,4. However, despite the explosive growth of generative artificial intelligence (AI), there have been few studies on building general-purpose multimodal AI assistants and copilots5 tailored to pathology. Here we present PathChat, a vision-language generalist AI assistant for human pathology. We built PathChat by adapting a foundational vision encoder for pathology, combining it with a pretrained large language model and fine-tuning the whole system on over 456,000 diverse visual-language instructions consisting of 999,202 question and answer turns. We compare PathChat with several multimodal vision-language AI assistants and GPT-4V, which powers the commercially available multimodal general-purpose AI assistant ChatGPT-4 (ref. 6). PathChat achieved state-of-the-art performance on multiple-choice diagnostic questions from cases with diverse tissue origins and disease models. Furthermore, using open-ended questions and human expert evaluation, we found that overall PathChat produced more accurate and pathologist-preferable responses to diverse queries related to pathology. As an interactive vision-language AI copilot that can flexibly handle both visual and natural language inputs, PathChat may potentially find impactful applications in pathology education, research and human-in-the-loop clinical decision-making.
© 2024. The Author(s).
Conflict of interest statement
A patent corresponding to this work has been filed by Mass General Brigham (Application 63/608,671). The tools, processes and models associated with PathChat have been exclusively licensed to ModellaAI. L.P.L., M.Y.L., R.J.C., B.C., F.M., D.F.K.W and J.J.W. hold equity interests in ModellaAI.
Figures









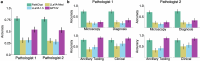

References
-
- Song, A. H. et al. Artificial intelligence for digital and computational pathology. Nat. Rev. Bioeng.1, 930–949 (2023).
-
- Shmatko, A. et al. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat. Cancer3, 1026–1038 (2022). - PubMed
-
- Ciga, O., Xu T. & Martel A. L. Self supervised contrastive learning for digital histopathology. Mach. Learn. Appl.7, 100198 (2022).
-
- Liu, H. et al. Visual instruction tuning. In Proc. Advances in Neural Information Processing Systems (eds Oh, A. et al.) 34892–34916 (Curran Associates, 2023).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

