Interactions between immune cell types facilitate the evolution of immune traits
- PMID: 38866051
- PMCID: PMC11306095
- DOI: 10.1038/s41586-024-07661-0
Interactions between immune cell types facilitate the evolution of immune traits
Abstract
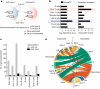
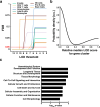
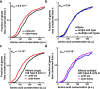
An essential prerequisite for evolution by natural selection is variation among individuals in traits that affect fitness1. The ability of a system to produce selectable variation, known as evolvability2, thus markedly affects the rate of evolution. Although the immune system is among the fastest-evolving components in mammals3, the sources of variation in immune traits remain largely unknown4,5. Here we show that an important determinant of the immune system's evolvability is its organization into interacting modules represented by different immune cell types. By profiling immune cell variation in bone marrow of 54 genetically diverse mouse strains from the Collaborative Cross6, we found that variation in immune cell frequencies is polygenic and that many associated genes are involved in homeostatic balance through cell-intrinsic functions of proliferation, migration and cell death. However, we also found genes associated with the frequency of a particular cell type that are expressed in a different cell type, exerting their effect in what we term cyto-trans. The vertebrate evolutionary record shows that genes associated in cyto-trans have faced weaker negative selection, thus increasing the robustness and hence evolvability2,7,8 of the immune system. This phenomenon is similarly observable in human blood. Our findings suggest that interactions between different components of the immune system provide a phenotypic space in which mutations can produce variation with little detriment, underscoring the role of modularity in the evolution of complex systems9.
© 2024. The Author(s).
Conflict of interest statement
S.S.O. holds equity and is a consultant of CytoReason and holds an unpaid position with the Human Immunome Project. T.D., E.S. and Y.A., are employees and hold equity in CytoReason. R.N holds equity in CytoReason.
Figures





Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Stigma Management Strategies of Autistic Social Media Users.Autism Adulthood. 2025 May 28;7(3):273-282. doi: 10.1089/aut.2023.0095. eCollection 2025 Jun. Autism Adulthood. 2025. PMID: 40539215
-
Maternal and neonatal outcomes of elective induction of labor.Evid Rep Technol Assess (Full Rep). 2009 Mar;(176):1-257. Evid Rep Technol Assess (Full Rep). 2009. PMID: 19408970 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
Cited by
-
Immuno-oncology recapitulates ontogeny: Modern cell and gene therapy for cancer.Mol Ther. 2025 May 7;33(5):2229-2237. doi: 10.1016/j.ymthe.2025.03.042. Epub 2025 Mar 27. Mol Ther. 2025. PMID: 40156188 Review.
-
Mapping the regulatory effects of common and rare non-coding variants across cellular and developmental contexts in the brain and heart.bioRxiv [Preprint]. 2025 Feb 20:2025.02.18.638922. doi: 10.1101/2025.02.18.638922. bioRxiv. 2025. PMID: 40027628 Free PMC article. Preprint.
-
The Human Cell Atlas from a cell census to a unified foundation model.Nature. 2025 Jan;637(8048):1065-1071. doi: 10.1038/s41586-024-08338-4. Epub 2024 Nov 20. Nature. 2025. PMID: 39566552
References
-
- Darwin, C. On the Origins of Species by Means of Natural Selection (Murray, 1859).
-
- Payne, J. L. & Wagner, A. The causes of evolvability and their evolution. Nat. Rev. Genet.20, 24–38 (2019). - PubMed
-
- Liston, A., Humblet-Baron, S., Duffy, D. & Goris, A. Human immune diversity: from evolution to modernity. Nat. Immunol.22, 1479–1489 (2021). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

