A nuclear architecture screen in Drosophila identifies Stonewall as a link between chromatin position at the nuclear periphery and germline stem cell fate
- PMID: 38866555
- PMCID: PMC11216176
- DOI: 10.1101/gad.351424.123
A nuclear architecture screen in Drosophila identifies Stonewall as a link between chromatin position at the nuclear periphery and germline stem cell fate
Abstract
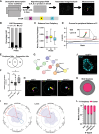
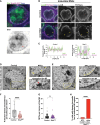
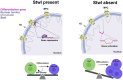
The association of genomic loci to the nuclear periphery is proposed to facilitate cell type-specific gene repression and influence cell fate decisions. However, the interplay between gene position and expression remains incompletely understood, in part because the proteins that position genomic loci at the nuclear periphery remain unidentified. Here, we used an Oligopaint-based HiDRO screen targeting ∼1000 genes to discover novel regulators of nuclear architecture in Drosophila cells. We identified the heterochromatin-associated protein Stonewall (Stwl) as a factor promoting perinuclear chromatin positioning. In female germline stem cells (GSCs), Stwl binds and positions chromatin loci, including GSC differentiation genes, at the nuclear periphery. Strikingly, Stwl-dependent perinuclear positioning is associated with transcriptional repression, highlighting a likely mechanism for Stwl's known role in GSC maintenance and ovary homeostasis. Thus, our study identifies perinuclear anchors in Drosophila and demonstrates the importance of gene repression at the nuclear periphery for cell fate.
Keywords: genome organization; germline stem cell; heterochromatin; nuclear architecture; nuclear periphery.
© 2024 Chavan et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Update of
-
A nuclear architecture screen in Drosophila identifies Stonewall as a link between chromatin position at the nuclear periphery and germline stem cell fate.bioRxiv [Preprint]. 2023 Nov 17:2023.11.17.567611. doi: 10.1101/2023.11.17.567611. bioRxiv. 2023. Update in: Genes Dev. 2024 Jun 25;38(9-10):415-435. doi: 10.1101/gad.351424.123. PMID: 38014085 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
