CircZNF609 and circNFIX as possible regulators of glioblastoma pathogenesis via miR-145-5p/EGFR axis
- PMID: 38866807
- PMCID: PMC11169398
- DOI: 10.1038/s41598-024-63827-w
CircZNF609 and circNFIX as possible regulators of glioblastoma pathogenesis via miR-145-5p/EGFR axis
Abstract
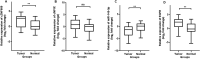
Glioblastoma is a rare and deadly malignancy with a low survival rate. Emerging evidence has shown that aberrantly expressed circular RNAs (circRNAs) play a critical role in the initiation and progression of GBM tumorigenesis. The oncogenic function of circZNF609 and circNFIX is involved in several types of cancer, but the role and underlying mechanism of these circRNAs in glioblastoma remain unclear. In this study, we hypothesized that circZNF609 and circNFIX may regulate EGFR through sponging miR-145-5p. Herein, we assessed the expression levels of circZNF609, circNFIX, miR-145-5p, and EGFR using quantitative polymerase chain reaction in glioblastoma patients and normal brain samples. The results showed that circZNF609, circNFIX, and EGFR expression levels were upregulated and miR145-5p was downregulated (p = 0.001, 0.06, 0.002, and 0.0065, respectively), while there was no significant association between clinicopathological features of the patients and the level of these genes expression. We also found a significant inverse correlation between miR145-5p and the expression of cZNF609, cNFIX and EGFR (p = 0.0003, 0.0006, and 0.009, respectively). These findings may open a new window for researchers to better understand the potential pathways involved in GBM pathogenesis. In conclusion, it may provide a new potential pathway for the development of effective drugs for the treatment of GBM patients.
Keywords: CircNFIX; CircZNF609; EGFR; miR-145-5p; Glioblastoma.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
Circular RNA ZNF609 promotes laryngeal squamous cell carcinoma progression by upregulating epidermal growth factor receptor via sponging microRNA-134-5p.Bioengineered. 2022 Mar;13(3):6929-6941. doi: 10.1080/21655979.2022.2034703. Bioengineered. 2022. PMID: 35236250 Free PMC article.
-
CircZNF609/miR-134-5p/BTG-2 axis regulates proliferation and migration of glioma cell.J Pharm Pharmacol. 2020 Jan;72(1):68-75. doi: 10.1111/jphp.13188. Epub 2019 Nov 13. J Pharm Pharmacol. 2020. PMID: 31721211
-
CircZNF609 enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the Hedgehog pathway through the regulation of miR-15a-5p/15b-5p and GLI2 expressions.Cell Death Dis. 2020 May 12;11(5):358. doi: 10.1038/s41419-020-2441-0. Cell Death Dis. 2020. PMID: 32398664 Free PMC article.
-
MicroRNAs involved in the EGFR pathway in glioblastoma.Biomed Pharmacother. 2021 Feb;134:111115. doi: 10.1016/j.biopha.2020.111115. Epub 2020 Dec 16. Biomed Pharmacother. 2021. PMID: 33341046 Review.
-
Exploring ncRNA-mediated regulation of EGFR signalling in glioblastoma: From mechanisms to therapeutics.Life Sci. 2024 May 15;345:122613. doi: 10.1016/j.lfs.2024.122613. Epub 2024 Apr 4. Life Sci. 2024. PMID: 38582393 Review.
Cited by
-
Hypoxic BMSC-derived exosomal miR-210-3p promotes progression of triple-negative breast cancer cells via NFIX-Wnt/β-catenin signaling axis.J Transl Med. 2025 Jan 9;23(1):39. doi: 10.1186/s12967-024-05947-5. J Transl Med. 2025. PMID: 39789572 Free PMC article.
References
-
- Wu M, et al. Circular RNAs: Characteristics, functions, mechanisms, and potential applications in thyroid cancer. Clin. Transl. Oncol. 2023;2023:1–17. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

