Induction and antiviral activity of ferret myxovirus resistance (Mx) protein 1 against influenza A viruses
- PMID: 38866913
- PMCID: PMC11169552
- DOI: 10.1038/s41598-024-63314-2
Induction and antiviral activity of ferret myxovirus resistance (Mx) protein 1 against influenza A viruses
Abstract
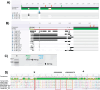
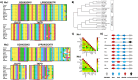
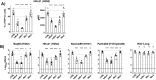
Myxovirus resistance (Mx) proteins are products of interferon stimulated genes (ISGs) and Mx proteins of different species have been reported to mediate antiviral activity against a number of viruses, including influenza A viruses (IAV). Ferrets are widely considered to represent the 'gold standard' small animal model for studying pathogenesis and immunity to human IAV infections, however little is known regarding the antiviral activity of ferret Mx proteins. Herein, we report induction of ferret (f)Mx1/2 in a ferret lung cell line and in airway tissues from IAV-infected ferrets, noting that fMx1 was induced to higher levels that fMx2 both in vitro and in vivo. Overexpression confirmed cytoplasmic expression of fMx1 as well as its ability to inhibit infection and replication of IAV, noting that this antiviral effect of fMx1was modest when compared to cells overexpressing either human MxA or mouse Mx1. Together, these studies provide the first insights regarding the role of fMx1 in cell innate antiviral immunity to influenza viruses. Understanding similarities and differences in the antiviral activities of human and ferret ISGs provides critical context for evaluating results when studying human IAV infections in the ferret model.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Induction of Interferon-Stimulated Genes Correlates with Reduced Growth of Influenza A Virus in Lungs after RIG-I Agonist Treatment of Ferrets.J Virol. 2022 Aug 24;96(16):e0055922. doi: 10.1128/jvi.00559-22. Epub 2022 Aug 2. J Virol. 2022. PMID: 35916513 Free PMC article.
-
Subcellular Localization of MxB Determines Its Antiviral Potential against Influenza A Virus.J Virol. 2020 Oct 27;94(22):e00125-20. doi: 10.1128/JVI.00125-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32907985 Free PMC article.
-
Delay of innate immune responses following influenza B virus infection affects the development of a robust antibody response in ferrets.mBio. 2025 Feb 5;16(2):e0236124. doi: 10.1128/mbio.02361-24. Epub 2025 Jan 8. mBio. 2025. PMID: 39772665 Free PMC article.
-
Mx genes: host determinants controlling influenza virus infection and trans-species transmission.Hum Genet. 2020 Jun;139(6-7):695-705. doi: 10.1007/s00439-019-02092-8. Epub 2019 Nov 26. Hum Genet. 2020. PMID: 31773252 Free PMC article. Review.
-
Mx GTPases: dynamin-like antiviral machines of innate immunity.Trends Microbiol. 2015 Mar;23(3):154-63. doi: 10.1016/j.tim.2014.12.003. Epub 2015 Jan 6. Trends Microbiol. 2015. PMID: 25572883 Review.
Cited by
-
Anthraquinone-2-Carboxylic Acid Is a Potential Antiviral Candidate Against Influenza Viruses In Vitro and In Vivo.Viruses. 2025 Apr 27;17(5):628. doi: 10.3390/v17050628. Viruses. 2025. PMID: 40431640 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

