The transcription factor PAX5 activates human LINE1 retrotransposons to induce cellular senescence
- PMID: 38866979
- PMCID: PMC11315925
- DOI: 10.1038/s44319-024-00176-9
The transcription factor PAX5 activates human LINE1 retrotransposons to induce cellular senescence
Abstract
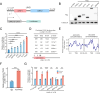
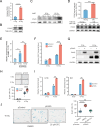
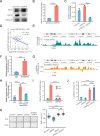
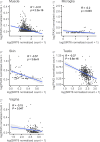
As a hallmark of senescent cells, the derepression of Long Interspersed Elements 1 (LINE1) transcription results in accumulated LINE1 cDNA, which triggers the secretion of the senescence-associated secretory phenotype (SASP) and paracrine senescence in a cGAS-STING pathway-dependent manner. However, transcription factors that govern senescence-associated LINE1 reactivation remain ill-defined. Here, we predict several transcription factors that bind to human LINE1 elements to regulate their transcription by analyzing the conserved binding motifs in the 5'-untranslated regions (UTR) of the commonly upregulated LINE1 elements in different types of senescent cells. Further analysis reveals that PAX5 directly binds to LINE1 5'-UTR and the binding is enhanced in senescent cells. The enrichment of PAX5 at the 5'-UTR promotes cellular senescence and SASP by activating LINE1. We also demonstrate that the longevity gene SIRT6 suppresses PAX5 transcription by directly binding to the PAX5 promoter, and overexpressing PAX5 abrogates the suppressive effect of SIRT6 on stress-dependent cellular senescence. Our work suggests that PAX5 could serve as a potential target for drug development aiming to suppress LINE1 activation and treat senescence-associated diseases.
Keywords: Cellular Senescence; LINE1; PAX5; SIRT6.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Bulut-Karslioglu A, De La Rosa-Velázquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galán C, Winter GE, Engist B, Gerle B et al (2014) Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell 55:277–290 10.1016/j.molcel.2014.05.029 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFA1103703/MOST | National Key Research and Development Program of China (NKPs)
- 2021YFA1102003/MOST | National Key Research and Development Program of China (NKPs)
- 2023M732662/China Postdoctoral Science Foundation (China Postdoctoral Foundation Project)
- 82225017/MOST | National Natural Science Foundation of China (NSFC)
- 32270750/MOST | National Natural Science Foundation of China (NSFC)
- 82071565/MOST | National Natural Science Foundation of China (NSFC)
- 32171288/MOST | National Natural Science Foundation of China (NSFC)
- 32200603/MOST | National Natural Science Foundation of China (NSFC)
- 32100605/MOST | National Natural Science Foundation of China (NSFC)
- 22YF1434500/Shanghai Sailing Program
- 21YF1436000/Shanghai Sailing Program
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

