Obesity induces PD-1 on macrophages to suppress anti-tumour immunity
- PMID: 38867043
- PMCID: PMC11456854
- DOI: 10.1038/s41586-024-07529-3
Obesity induces PD-1 on macrophages to suppress anti-tumour immunity
Erratum in
-
Author Correction: Obesity induces PD-1 on macrophages to suppress anti-tumour immunity.Nature. 2024 Jul;631(8022):E16. doi: 10.1038/s41586-024-07794-2. Nature. 2024. PMID: 38982216 No abstract available.
Abstract
Obesity is a leading risk factor for progression and metastasis of many cancers1,2, yet can in some cases enhance survival3-5 and responses to immune checkpoint blockade therapies, including anti-PD-1, which targets PD-1 (encoded by PDCD1), an inhibitory receptor expressed on immune cells6-8. Although obesity promotes chronic inflammation, the role of the immune system in the obesity-cancer connection and immunotherapy remains unclear. It has been shown that in addition to T cells, macrophages can express PD-19-12. Here we found that obesity selectively induced PD-1 expression on tumour-associated macrophages (TAMs). Type I inflammatory cytokines and molecules linked to obesity, including interferon-γ, tumour necrosis factor, leptin, insulin and palmitate, induced macrophage PD-1 expression in an mTORC1- and glycolysis-dependent manner. PD-1 then provided negative feedback to TAMs that suppressed glycolysis, phagocytosis and T cell stimulatory potential. Conversely, PD-1 blockade increased the level of macrophage glycolysis, which was essential for PD-1 inhibition to augment TAM expression of CD86 and major histocompatibility complex I and II molecules and ability to activate T cells. Myeloid-specific PD-1 deficiency slowed tumour growth, enhanced TAM glycolysis and antigen-presentation capability, and led to increased CD8+ T cell activity with a reduced level of markers of exhaustion. These findings show that obesity-associated metabolic signalling and inflammatory cues cause TAMs to induce PD-1 expression, which then drives a TAM-specific feedback mechanism that impairs tumour immune surveillance. This may contribute to increased cancer risk yet improved response to PD-1 immunotherapy in obesity.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures
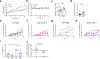




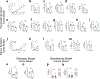


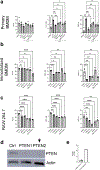
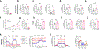



Comment in
-
Macrophages dig into the obesity paradox in cancer.Immunity. 2024 Aug 13;57(8):1731-1733. doi: 10.1016/j.immuni.2024.07.013. Immunity. 2024. PMID: 39142274
References
-
- Sung H et al. Global patterns in excess body weight and the associated cancer burden. CA Cancer J. Clin. 69, 88–112 (2019). - PubMed
-
- Choi Y et al. Body mass index and survival in patients with renal cell carcinoma: a clinical-based cohort and meta-analysis. Int. J. Cancer 132, 625–634 (2013). - PubMed
-
- Schlesinger S et al. Postdiagnosis body mass index and risk of mortality in colorectal cancer survivors: a prospective study and meta-analysis. Cancer Causes Control 25, 1407–1418 (2014). - PubMed
MeSH terms
Substances
Grants and funding
- K00 CA234920/CA/NCI NIH HHS/United States
- U01 CA272541/CA/NCI NIH HHS/United States
- K12 CA090625/CA/NCI NIH HHS/United States
- R01 DK105550/DK/NIDDK NIH HHS/United States
- G20 RR030956/RR/NCRR NIH HHS/United States
- T32 DK101003/DK/NIDDK NIH HHS/United States
- T32 GM007347/GM/NIGMS NIH HHS/United States
- F99 CA274695/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- K08 CA241351/CA/NCI NIH HHS/United States
- T32 DK007314/DK/NIDDK NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- K00 CA253718/CA/NCI NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- F30 CA239367/CA/NCI NIH HHS/United States
- F31 CA261049/CA/NCI NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- R01 CA238263/CA/NCI NIH HHS/United States
- F30 CA247202/CA/NCI NIH HHS/United States
- R01 CA217987/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

