CTLA-4-expressing ILC3s restrain interleukin-23-mediated inflammation
- PMID: 38867048
- PMCID: PMC11298788
- DOI: 10.1038/s41586-024-07537-3
CTLA-4-expressing ILC3s restrain interleukin-23-mediated inflammation
Abstract
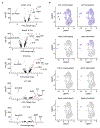
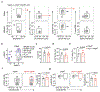
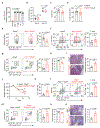
Interleukin (IL-)23 is a major mediator and therapeutic target in chronic inflammatory diseases that also elicits tissue protection in the intestine at homeostasis or following acute infection1-4. However, the mechanisms that shape these beneficial versus pathological outcomes remain poorly understood. To address this gap in knowledge, we performed single-cell RNA sequencing on all IL-23 receptor-expressing cells in the intestine and their acute response to IL-23, revealing a dominance of T cells and group 3 innate lymphoid cells (ILC3s). Unexpectedly, we identified potent upregulation of the immunoregulatory checkpoint molecule cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) on ILC3s. This pathway was activated by gut microbes and IL-23 in a FOXO1- and STAT3-dependent manner. Mice lacking CTLA-4 on ILC3s exhibited reduced regulatory T cells, elevated inflammatory T cells and more-severe intestinal inflammation. IL-23 induction of CTLA-4+ ILC3s was necessary and sufficient to reduce co-stimulatory molecules and increase PD-L1 bioavailability on intestinal myeloid cells. Finally, human ILC3s upregulated CTLA-4 in response to IL-23 or gut inflammation and correlated with immunoregulation in inflammatory bowel disease. These results reveal ILC3-intrinsic CTLA-4 as an essential checkpoint that restrains the pathological outcomes of IL-23, suggesting that disruption of these lymphocytes, which occurs in inflammatory bowel disease5-7, contributes to chronic inflammation.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures







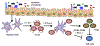




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

