PRDX6 augments selenium utilization to limit iron toxicity and ferroptosis
- PMID: 38867112
- PMCID: PMC11327102
- DOI: 10.1038/s41594-024-01329-z
PRDX6 augments selenium utilization to limit iron toxicity and ferroptosis
Abstract
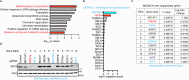
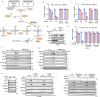
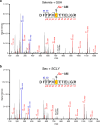
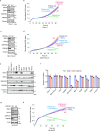
Ferroptosis is a form of regulated cell death induced by iron-dependent accumulation of lipid hydroperoxides. Selenoprotein glutathione peroxidase 4 (GPX4) suppresses ferroptosis by detoxifying lipid hydroperoxides via a catalytic selenocysteine (Sec) residue. Sec, the genetically encoded 21st amino acid, is biosynthesized from a reactive selenium donor on its cognate tRNA[Ser]Sec. It is thought that intracellular selenium must be delivered 'safely' and 'efficiently' by a carrier protein owing to its high reactivity and very low concentrations. Here, we identified peroxiredoxin 6 (PRDX6) as a novel selenoprotein synthesis factor. Loss of PRDX6 decreases the expression of selenoproteins and induces ferroptosis via a reduction in GPX4. Mechanistically, PRDX6 increases the efficiency of intracellular selenium utilization by transferring selenium between proteins within the selenocysteyl-tRNA[Ser]Sec synthesis machinery, leading to efficient synthesis of selenocysteyl-tRNA[Ser]Sec. These findings highlight previously unidentified selenium metabolic systems and provide new insights into ferroptosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

