APOE2 protects against Aβ pathology by improving neuronal mitochondrial function through ERRα signaling
- PMID: 38867189
- PMCID: PMC11170814
- DOI: 10.1186/s11658-024-00600-x
APOE2 protects against Aβ pathology by improving neuronal mitochondrial function through ERRα signaling
Abstract
Background: Alzheimer's disease (AD) is a progressive neurodegenerative disease and apolipoprotein E (APOE) genotypes (APOE2, APOE3, and APOE4) show different AD susceptibility. Previous studies indicated that individuals carrying the APOE2 allele reduce the risk of developing AD, which may be attributed to the potential neuroprotective role of APOE2. However, the mechanisms underlying the protective effects of APOE2 is still unclear.
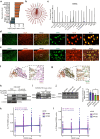
Methods: We analyzed single-nucleus RNA sequencing and bulk RNA sequencing data of APOE2 and APOE3 carriers from the Religious Orders Study and Memory and Aging Project (ROSMAP) cohort. We validated the findings in SH-SY5Y cells and AD model mice by evaluating mitochondrial functions and cognitive behaviors respectively.
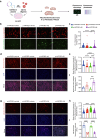
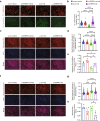
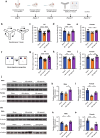
Results: The pathway analysis of six major cell types revealed a strong association between APOE2 and cellular stress and energy metabolism, particularly in excitatory and inhibitory neurons, which was found to be more pronounced in the presence of beta-amyloid (Aβ). Moreover, APOE2 overexpression alleviates Aβ1-42-induced mitochondrial dysfunction and reduces the generation of reactive oxygen species in SH-SY5Y cells. These protective effects may be due to ApoE2 interacting with estrogen-related receptor alpha (ERRα). ERRα overexpression by plasmids or activation by agonist was also found to show similar mitochondrial protective effects in Aβ1-42-stimulated SH-SY5Y cells. Additionally, ERRα agonist treatment improve the cognitive performance of Aβ injected mice in both Y maze and novel object recognition tests. ERRα agonist treatment increased PSD95 expression in the cortex of agonist-treated-AD mice.
Conclusions: APOE2 appears to enhance neural mitochondrial function via the activation of ERRα signaling, which may be the protective effect of APOE2 to treat AD.
Keywords: Alzheimer's disease; Apolipoprotein E; Beta-amyloid (Aβ); ESRRA; Mitochondria; Neuron.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Differential Signaling Mediated by ApoE2, ApoE3, and ApoE4 in Human Neurons Parallels Alzheimer's Disease Risk.J Neurosci. 2019 Sep 11;39(37):7408-7427. doi: 10.1523/JNEUROSCI.2994-18.2019. Epub 2019 Jul 22. J Neurosci. 2019. PMID: 31331998 Free PMC article.
-
Opposing effects of viral mediated brain expression of apolipoprotein E2 (apoE2) and apoE4 on apoE lipidation and Aβ metabolism in apoE4-targeted replacement mice.Mol Neurodegener. 2015 Mar 5;10:6. doi: 10.1186/s13024-015-0001-3. Mol Neurodegener. 2015. PMID: 25871773 Free PMC article.
-
Blocking the apoE/Aβ interaction ameliorates Aβ-related pathology in APOE ε2 and ε4 targeted replacement Alzheimer model mice.Acta Neuropathol Commun. 2014 Jun 28;2:75. doi: 10.1186/s40478-014-0075-0. Acta Neuropathol Commun. 2014. PMID: 24972680 Free PMC article.
-
Apolipoprotein E and oxidative stress in brain with relevance to Alzheimer's disease.Neurobiol Dis. 2020 May;138:104795. doi: 10.1016/j.nbd.2020.104795. Epub 2020 Feb 6. Neurobiol Dis. 2020. PMID: 32036033 Free PMC article. Review.
-
Apolipoprotein E isoforms in Alzheimer's disease pathology and etiology.Microsc Res Tech. 2000 Aug 15;50(4):278-81. doi: 10.1002/1097-0029(20000815)50:4<278::AID-JEMT5>3.0.CO;2-T. Microsc Res Tech. 2000. PMID: 10936880 Review.
Cited by
-
Integrating neuroprotection, antioxidative effects, and precision medicine in glaucoma management with bioactive compounds.Biomed Pharmacother. 2025 Sep;190:118319. doi: 10.1016/j.biopha.2025.118319. Epub 2025 Jul 18. Biomed Pharmacother. 2025. PMID: 40683209 Free PMC article. Review.
-
Smelling the Risk: Early Olfactory Deficits, Brain Networks, and Blood Markers of Alzheimer's Disease Risk in Humanized APOE Mice.bioRxiv [Preprint]. 2025 Jun 9:2025.06.06.658383. doi: 10.1101/2025.06.06.658383. bioRxiv. 2025. PMID: 40661485 Free PMC article. Preprint.
-
Olfactory-Guided Behavior Uncovers Imaging and Molecular Signatures of Alzheimer's Disease Risk.Brain Sci. 2025 Aug 13;15(8):863. doi: 10.3390/brainsci15080863. Brain Sci. 2025. PMID: 40867194 Free PMC article.
-
Proteomic signatures of the APOE ε4 and APOE ε2 genetic variants and Alzheimer's disease.medRxiv [Preprint]. 2025 Aug 25:2025.08.04.25332945. doi: 10.1101/2025.08.04.25332945. medRxiv. 2025. PMID: 40799961 Free PMC article. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

