Exploring the microbiota difference of bronchoalveolar lavage fluid between community-acquired pneumonia with or without COPD based on metagenomic sequencing: a retrospective study
- PMID: 38867204
- PMCID: PMC11167785
- DOI: 10.1186/s12890-024-03087-6
Exploring the microbiota difference of bronchoalveolar lavage fluid between community-acquired pneumonia with or without COPD based on metagenomic sequencing: a retrospective study
Abstract
Background: Community-acquired pneumonia (CAP) patients with chronic obstructive pulmonary disease (COPD) have higher disease severity and mortality compared to those without COPD. However, deep investigation into microbiome distribution of lower respiratory tract of CAP with or without COPD was unknown.
Methods: So we used metagenomic next generation sequencing (mNGS) to explore the microbiome differences between the two groups.
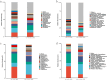
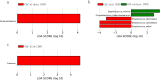
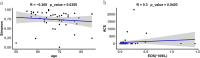
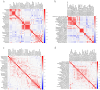
Results: Thirty-six CAP without COPD and 11 CAP with COPD cases were retrieved. Bronchoalveolar lavage fluid (BALF) was collected and analyzed using untargeted mNGS and bioinformatic analysis. mNGS revealed that CAP with COPD group was abundant with Streptococcus, Prevotella, Bordetella at genus level and Cutibacterium acnes, Rothia mucilaginosa, Bordetella genomosp. 6 at species level. While CAP without COPD group was abundant with Ralstonia, Prevotella, Streptococcus at genus level and Ralstonia pickettii, Rothia mucilaginosa, Prevotella melaninogenica at species level. Meanwhile, both alpha and beta microbiome diversity was similar between groups. Linear discriminant analysis found that pa-raburkholderia, corynebacterium tuberculostearicum and staphylococcus hominis were more enriched in CAP without COPD group while the abundance of streptococcus intermedius, streptococcus constellatus, streptococcus milleri, fusarium was higher in CAP with COPD group.
Conclusions: These findings revealed that concomitant COPD have an mild impact on lower airway microbiome of CAP patients.
Keywords: Bronchoalveolar lavage fluid; Chronic obstructive pulmonary disease; Community acquired pneumonia; Metagenomic next-generation sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
A metagenomic next-generation sequencing (mNGS)-based analysis of bronchoalveolar lavage samples in patients with an acute exacerbation of chronic obstructive pulmonary disease.J Mol Histol. 2024 Oct;55(5):709-719. doi: 10.1007/s10735-024-10225-1. Epub 2024 Jul 26. J Mol Histol. 2024. PMID: 39060894
-
The etiological diagnostic value of metagenomic next-generation sequencing in suspected community-acquired pneumonia.BMC Infect Dis. 2024 Jun 24;24(1):626. doi: 10.1186/s12879-024-09507-6. BMC Infect Dis. 2024. PMID: 38914949 Free PMC article.
-
Analysis of the bronchoalveolar lavage fluid microbial flora in COPD patients at different lung function during acute exacerbation.Sci Rep. 2025 Apr 16;15(1):13179. doi: 10.1038/s41598-025-96746-5. Sci Rep. 2025. PMID: 40240456 Free PMC article.
-
Current Status of Community-Acquired Pneumonia in Patients with Chronic Obstructive Pulmonary Disease.Chin Med J (Engl). 2018 May 5;131(9):1086-1091. doi: 10.4103/0366-6999.230727. Chin Med J (Engl). 2018. PMID: 29692381 Free PMC article. Review.
-
Metagenomics next-generation sequencing tests take the stage in the diagnosis of lower respiratory tract infections.J Adv Res. 2021 Sep 29;38:201-212. doi: 10.1016/j.jare.2021.09.012. eCollection 2022 May. J Adv Res. 2021. PMID: 35572406 Free PMC article. Review.
Cited by
-
Microbial profiling of community-acquired pneumonia in patients with and without chronic obstructive pulmonary disease: a comprehensive molecular diagnostics study.Pneumonia (Nathan). 2025 Aug 5;17(1):19. doi: 10.1186/s41479-025-00172-0. Pneumonia (Nathan). 2025. PMID: 40760674 Free PMC article.
-
Investigating the Role of Invasive Streptococcus Constellatus Infection in Severe Systemic Disease Manifestations: A Case Report.Cureus. 2024 Aug 17;16(8):e67088. doi: 10.7759/cureus.67088. eCollection 2024 Aug. Cureus. 2024. PMID: 39286694 Free PMC article.
-
Microbial and clinical disparities in pneumonia: insights from metagenomic next-generation sequencing in patients with community-acquired and severe pneumonia.Front Microbiol. 2025 Jun 20;16:1538109. doi: 10.3389/fmicb.2025.1538109. eCollection 2025. Front Microbiol. 2025. PMID: 40620487 Free PMC article.
References
-
- Jiang N, Long QY, Zheng YL, Gao ZC. Advances in epidemiology, etiology, and treatment of community-acquired pneumonia. Zhonghua yu fang yi xue za zhi. 2023;57(1):91–99. - PubMed
-
- Hsu CW, Suk CW, Hsu YP, Chang JH, Liu CT, Huang SK, Hsu SC. Sphingosine-1-phosphate and CRP as potential combination biomarkers in discrimination of COPD with community-acquired pneumonia and acute exacerbation of COPD. Respir Res. 2022;23(1):63. doi: 10.1186/s12931-022-01991-1. - DOI - PMC - PubMed
-
- Yang J, Zhang Q, Zhang J, Ouyang Y, Sun Z, Liu X, Qaio F, Xu LQ, Niu Y, Li J. Exploring the Change of Host and Microorganism in Chronic Obstructive Pulmonary Disease Patients Based on Metagenomic and Metatranscriptomic Sequencing. Front Microbiol. 2022;13:818281. doi: 10.3389/fmicb.2022.818281. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

