Comparative transcriptome analysis reveals molecular damage associated with cryopreservation in Crassostrea angulata D-larvae rather than to cryoprotectant exposure
- PMID: 38867206
- PMCID: PMC11167747
- DOI: 10.1186/s12864-024-10473-1
Comparative transcriptome analysis reveals molecular damage associated with cryopreservation in Crassostrea angulata D-larvae rather than to cryoprotectant exposure
Abstract
Background: The Portuguese oyster Crassostrea angulata, a bivalve of significant economic and ecological importance, has faced a decline in both production and natural populations due to pathologies, climate change, and anthropogenic factors. To safeguard its genetic diversity and improve reproductive management, cryopreservation emerges as a valuable strategy. However, the cryopreservation methodologies lead to some damage in structures and functions of the cells and tissues that can affect post-thaw quality. Transcriptomics may help to understand the molecular consequences related to cryopreservation steps and therefore to identify different freezability biomarkers. This study investigates the molecular damage induced by cryopreservation in C. angulata D-larvae, focusing on two critical steps: exposure to cryoprotectant solution and the freezing/thawing process.
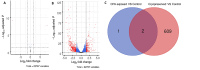
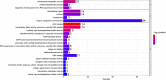
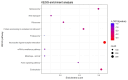
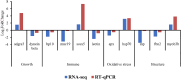
Results: Expression analysis revealed 3 differentially expressed genes between larvae exposed to cryoprotectant solution and fresh larvae and 611 differentially expressed genes in cryopreserved larvae against fresh larvae. The most significantly enriched gene ontology terms were "carbohydrate metabolic process", "integral component of membrane" and "chitin binding" for biological processes, cellular components and molecular functions, respectively. Kyoto Encyclopedia of Genes and Genomes enrichment analysis identified the "neuroactive ligand receptor interaction", "endocytosis" and "spliceosome" as the most enriched pathways. RNA sequencing results were validate by quantitative RT-PCR, once both techniques presented the same gene expression tendency and a group of 11 genes were considered important molecular biomarkers to be used in further studies for the evaluation of cryodamage.
Conclusions: The current work provided valuable insights into the molecular repercussions of cryopreservation on D-larvae of Crassostrea angulata, revealing that the freezing process had a more pronounced impact on larval quality compared to any potential cryoprotectant-induced toxicity. Additionally, was identify 11 genes serving as biomarkers of freezability for D-larvae quality assessment. This research contributes to the development of more effective cryopreservation protocols and detection methods for cryodamage in this species.
Keywords: Cryodamage; Cryoprotectant exposure; D-larvae cryopreservation; Gene expression; Portuguese oyster; RNA-seq.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Wang H, Qian L, Liu X, Zhang G, Guo X. Classification of a common cupped oyster from Southern China. J Shellfish Res. 2010;29:857–66. doi: 10.2983/035.029.0420. - DOI
-
- Grizel H, Héral M. Introduction into France of the Japanese oyster (Crassostrea gigas) ICES J Mar Sci. 1991;47:399–403. doi: 10.1093/icesjms/47.3.399. - DOI
-
- Huvet A, Lapègue S, Magoulas A, Boudry P. Mitochondrial and nuclear DNA phylogeography of Crassostrea angulata, the Portuguese oyster endangered in Europe. Conserv Genet. 2000;1:251–62. doi: 10.1023/A:1011505805923. - DOI
-
- Anjos C, Baptista T, Joaquim S, Mendes S, Matias AM, Moura P, et al. Broodstock conditioning of the Portuguese oyster (Crassostrea angulata, Lamarck, 1819): influence of different diets. Aquac Res. 2017;48:3859–78. doi: 10.1111/are.13213. - DOI
-
- Adams SL, Smith JF, Roberts RD, Janke AR, King NG, Tervit HR, et al. Application of sperm cryopreservation in selective breeding of the Pacific oyster, Crassostrea gigas (Thunberg) Aquac Res. 2008;39:1434–42. doi: 10.1111/j.1365-2109.2008.02013.x. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

