A DIO2 missense mutation and its impact on fetal response to PRRSV infection
- PMID: 38867209
- PMCID: PMC11167750
- DOI: 10.1186/s12917-024-04099-4
A DIO2 missense mutation and its impact on fetal response to PRRSV infection
Abstract
Background: Porcine reproductive and respiratory syndrome virus 2 (PRRSV-2) infection during late gestation substantially lowers fetal viability and survival. In a previous genome-wide association study, a single nucleotide polymorphism on chromosome 7 was significantly associated with probability of fetuses being viable in response to maternal PRRSV-2 infection at 21 days post maternal inoculation. The iodothyronine deiodinase 2 (DIO2) gene, located ~ 14 Kilobase downstream of this SNP, was selected as a priority candidate related to fetal susceptibility following maternal PRRSV-2 infection. Our objectives were to identify mutation(s) within the porcine DIO2 gene and to determine if they were associated with fetal outcomes after PRRSV-2 challenge. Sequencing of the DIO2, genotyping identified variants, and association of DIO2 genotypes with fetal phenotypes including DIO2 mRNA levels, viability, survival, viral loads, cortisol and thyroid hormone levels, and growth measurements were conducted.
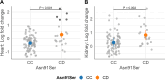
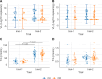
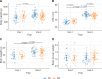
Results: A missense variant (p.Asn91Ser) was identified in the parental populations from two independent PRRSV-2 challenge trials. This variant was further genotyped to determine association with fetal PRRS outcomes. DIO2 mRNA levels in fetal heart and kidney differed by the genotypes of Asn91Ser substitution with significantly greater DIO2 mRNA expression in heterozygotes compared with wild-type homozygotes (P < 0.001 for heart, P = 0.002 for kidney). While Asn91Ser did not significantly alter fetal viability and growth measurements, interaction effects of the variant with fetal sex or trial were identified for fetal viability or crown rump length, respectively. However, this mutation was not related to dysregulation of the hypothalamic-pituitary-adrenal and thyroid axis, indicated by no differences in circulating cortisol, T4, and T3 levels in fetuses of the opposing genotypes following PRRSV-2 infection.
Conclusions: The present study suggests that a complex relationship among DIO2 genotype, DIO2 expression, fetal sex, and fetal viability may exist during the course of fetal PRRSV infection. Our study also proposes the increase in cortisol levels, indicative of fetal stress response, may lead to fetal complications, such as fetal compromise, fetal death, or premature farrowing, during PRRSV infection.
Keywords: Deiodinase; Fetus; Nonsynonymous mutation; PRRSV; Swine; Thyroid.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Effects of porcine reproductive and respiratory syndrome virus (PRRSV) on thyroid hormone metabolism in the late gestation fetus.Vet Res. 2022 Sep 30;53(1):74. doi: 10.1186/s13567-022-01092-3. Vet Res. 2022. PMID: 36175938 Free PMC article.
-
Variation in fetal outcome, viral load and ORF5 sequence mutations in a large scale study of phenotypic responses to late gestation exposure to type 2 porcine reproductive and respiratory syndrome virus.PLoS One. 2014 Apr 22;9(4):e96104. doi: 10.1371/journal.pone.0096104. eCollection 2014. PLoS One. 2014. PMID: 24756023 Free PMC article.
-
Classification of fetal resilience to porcine reproductive and respiratory syndrome (PRRS) based on temporal viral load in late gestation maternal tissues and fetuses.Virus Res. 2019 Jan 15;260:151-162. doi: 10.1016/j.virusres.2018.12.002. Epub 2018 Dec 5. Virus Res. 2019. PMID: 30529234
-
Novel insights into host responses and reproductive pathophysiology of porcine reproductive and respiratory syndrome caused by PRRSV-2.Vet Microbiol. 2017 Sep;209:114-123. doi: 10.1016/j.vetmic.2017.02.019. Epub 2017 Mar 2. Vet Microbiol. 2017. PMID: 28292546 Review.
-
Porcine reproductive and respiratory syndrome.Vet Pathol. 1998 Jan;35(1):1-20. doi: 10.1177/030098589803500101. Vet Pathol. 1998. PMID: 9545131 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

