Digital in-line holographic microscopy for label-free identification and tracking of biological cells
- PMID: 38867274
- PMCID: PMC11170804
- DOI: 10.1186/s40779-024-00541-8
Digital in-line holographic microscopy for label-free identification and tracking of biological cells
Abstract
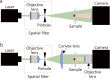
Digital in-line holographic microscopy (DIHM) is a non-invasive, real-time, label-free technique that captures three-dimensional (3D) positional, orientational, and morphological information from digital holographic images of living biological cells. Unlike conventional microscopies, the DIHM technique enables precise measurements of dynamic behaviors exhibited by living cells within a 3D volume. This review outlines the fundamental principles and comprehensive digital image processing procedures employed in DIHM-based cell tracking methods. In addition, recent applications of DIHM technique for label-free identification and digital tracking of various motile biological cells, including human blood cells, spermatozoa, diseased cells, and unicellular microorganisms, are thoroughly examined. Leveraging artificial intelligence has significantly enhanced both the speed and accuracy of digital image processing for cell tracking and identification. The quantitative data on cell morphology and dynamics captured by DIHM can effectively elucidate the underlying mechanisms governing various microbial behaviors and contribute to the accumulation of diagnostic databases and the development of clinical treatments.
Keywords: Artificial intelligence; Cell identification; Cell tracking; Digital in-line holographic microscopy (DIHM).
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Cierpka C, Kähler CJ. Particle imaging techniques for volumetric three-component (3D3C) velocity measurements in microfluidics. J Vis. 2012;15:1–31. doi: 10.1007/s12650-011-0107-9. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

