Claudin-4 Modulates Autophagy via SLC1A5/LAT1 as a Mechanism to Regulate Micronuclei
- PMID: 38867360
- PMCID: PMC11218812
- DOI: 10.1158/2767-9764.CRC-24-0240
Claudin-4 Modulates Autophagy via SLC1A5/LAT1 as a Mechanism to Regulate Micronuclei
Abstract
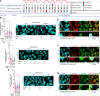
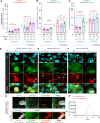
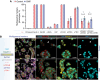
Genome instability is a hallmark of cancer crucial for tumor heterogeneity and is often a result of defects in cell division and DNA damage repair. Tumors tolerate genomic instability, but the accumulation of genetic aberrations is regulated to avoid catastrophic chromosomal alterations and cell death. In ovarian cancer tumors, claudin-4 is frequently upregulated and closely associated with genome instability and worse patient outcomes. However, its biological association with regulating genomic instability is poorly understood. Here, we used CRISPR interference and a claudin mimic peptide to modulate the claudin-4 expression and its function in vitro and in vivo. We found that claudin-4 promotes a tolerance mechanism for genomic instability through micronuclei generation in tumor cells. Disruption of claudin-4 increased autophagy and was associated with the engulfment of cytoplasm-localized DNA. Mechanistically, we observed that claudin-4 establishes a biological axis with the amino acid transporters SLC1A5 and LAT1, which regulate autophagy upstream of mTOR. Furthermore, the claudin-4/SLC1A5/LAT1 axis was linked to the transport of amino acids across the plasma membrane as one of the potential cellular processes that significantly decreased survival in ovarian cancer patients. Together, our results show that the upregulation of claudin-4 contributes to increasing the threshold of tolerance for genomic instability in ovarian tumor cells by limiting its accumulation through autophagy.
Significance: Autophagy regulation via claudin-4/SLC1A5/LAT1 has the potential to be a targetable mechanism to interfere with genomic instability in ovarian tumor cells.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. J. Lang reports grants from DOD during the conduct of the study. B.G. Bitler reports grants from NCI/NIH, ACS, DOD, and OCRA during the conduct of the study. No other disclosures were reported.
Figures
Similar articles
-
Claudin-4 Stabilizes the Genome via Nuclear and Cell-Cycle Remodeling to Support Ovarian Cancer Cell Survival.Cancer Res Commun. 2025 Jan 1;5(1):39-53. doi: 10.1158/2767-9764.CRC-24-0558. Cancer Res Commun. 2025. PMID: 39625235 Free PMC article.
-
Claudin-4 modulates autophagy via SLC1A5/LAT1 as a tolerance mechanism for genomic instability in ovarian cancer.bioRxiv [Preprint]. 2024 Jan 22:2024.01.18.576263. doi: 10.1101/2024.01.18.576263. bioRxiv. 2024. PMID: 38293054 Free PMC article. Preprint.
-
Claudin-4 activity in ovarian tumor cell apoptosis resistance and migration.BMC Cancer. 2016 Oct 11;16(1):788. doi: 10.1186/s12885-016-2799-7. BMC Cancer. 2016. PMID: 27724921 Free PMC article.
-
Potential roles of claudin-3 and claudin-4 in ovarian cancer management.J Egypt Natl Canc Inst. 2022 Jun 6;34(1):24. doi: 10.1186/s43046-022-00125-4. J Egypt Natl Canc Inst. 2022. PMID: 35665865 Review.
-
Claudins overexpression in ovarian cancer: potential targets for Clostridium Perfringens Enterotoxin (CPE) based diagnosis and therapy.Int J Mol Sci. 2013 May 17;14(5):10412-37. doi: 10.3390/ijms140510412. Int J Mol Sci. 2013. PMID: 23685873 Free PMC article. Review.
Cited by
-
Claudin-4 Stabilizes the Genome via Nuclear and Cell-Cycle Remodeling to Support Ovarian Cancer Cell Survival.Cancer Res Commun. 2025 Jan 1;5(1):39-53. doi: 10.1158/2767-9764.CRC-24-0558. Cancer Res Commun. 2025. PMID: 39625235 Free PMC article.
-
Extrajunctional CLDN10 cooperates with LAT1 and accelerates clear cell renal cell carcinoma progression.Cell Commun Signal. 2024 Dec 5;22(1):588. doi: 10.1186/s12964-024-01964-5. Cell Commun Signal. 2024. PMID: 39639312 Free PMC article.
-
Claudin-4 remodeling of nucleus-cell cycle crosstalk maintains ovarian tumor genome stability and drives resistance to genomic instability-inducing agents.bioRxiv [Preprint]. 2024 Sep 7:2024.09.04.611120. doi: 10.1101/2024.09.04.611120. bioRxiv. 2024. PMID: 39282307 Free PMC article. Preprint.
-
Role of solute carrier transporters in ovarian cancer (Review).Int J Mol Med. 2025 Feb;55(2):24. doi: 10.3892/ijmm.2024.5465. Epub 2024 Nov 29. Int J Mol Med. 2025. PMID: 39611477 Free PMC article. Review.
References
-
- Ovarian Cancer Research Alliance . Ovarian cancer statistics; 2024. [cited Dec 1 2024].
-
- Arora T, Mullangi S, Lekkala MR. Ovarian cancer. Treasure Island (FL): StatPearls2023; 2024.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

