Integrated Single-Cell RNA-seq and ATAC-seq Reveals Heterogeneous Differentiation of CD4+ Naive T Cell Subsets is Associated with Response to Antidepressant Treatment in Major Depressive Disorder
- PMID: 38867657
- PMCID: PMC11321657
- DOI: 10.1002/advs.202308393
Integrated Single-Cell RNA-seq and ATAC-seq Reveals Heterogeneous Differentiation of CD4+ Naive T Cell Subsets is Associated with Response to Antidepressant Treatment in Major Depressive Disorder
Abstract
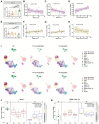
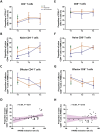
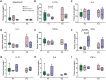
The mechanism involved in major depressive disorder (MDD) is well-studied but the mechanistic origin of the heterogeneous antidepressant effect remains largely unknown. Single-cell RNA-sequencing (scRNA-seq) and assay for transposase-accessible chromatin using sequencing (ATAC-seq) on peripheral blood mononuclear cells from 8 healthy individuals and 8 MDD patients before or after 12 weeks of antidepressant treatment is performed. scRNA-seq analysis reveals a lower proportion of naive T cells, particularly CD4+ naive T cells, in MDD patients compared to controls, and in nonresponders versus responders at the baseline. Flow cytometry data analysis of an independent cohort of 35 patients and 40 healthy individuals confirms the findings. Enrichment analysis of differentially expressed genes indicated obvious immune activation in responders. A specific activated CD4+ naive T population in responders characterized by enhanced mitogen-activated protein kinases (MAPK) pathway is identified. E-twenty six (ETS) is proposed as an upstream regulator of the MAPK pathway and heterogeneous differentiation in activated CD4+ naive T population is associated with the response to antidepressant treatment in MDD patients. A distinct immune feature manifested by CD4+ naive T cells during antidepressant treatment in MDD is identified. Collectively, this proposes the molecular mechanism that underlies the heterogeneous antidepressant outcomes for MDD.
Keywords: ATAC sequencing; CD4+ naive T cells; major depressive disorder; single‐cell RNA sequencing.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Repertoire-based mapping and time-tracking of T helper cell subsets in scRNA-Seq.Front Immunol. 2025 Apr 4;16:1536302. doi: 10.3389/fimmu.2025.1536302. eCollection 2025. Front Immunol. 2025. PMID: 40255395 Free PMC article.
-
Circulating cytotoxic T cells and natural killer cells as potential predictors for antidepressant response in melancholic depression. Restoration of T regulatory cell populations after antidepressant therapy.Psychopharmacology (Berl). 2016 May;233(9):1679-88. doi: 10.1007/s00213-015-3943-9. Epub 2015 May 8. Psychopharmacology (Berl). 2016. PMID: 25953327
-
Epigenetic Landscapes of Single-Cell Chromatin Accessibility and Transcriptomic Immune Profiles of T Cells in COVID-19 Patients.Front Immunol. 2021 Feb 24;12:625881. doi: 10.3389/fimmu.2021.625881. eCollection 2021. Front Immunol. 2021. PMID: 33717140 Free PMC article.
-
Application of computational algorithms for single-cell RNA-seq and ATAC-seq in neurodegenerative diseases.Brief Funct Genomics. 2025 Jan 15;24:elae044. doi: 10.1093/bfgp/elae044. Brief Funct Genomics. 2025. PMID: 39500613 Free PMC article. Review.
-
Peripheral CD4+ T helper lymphocytes alterations in major depressive disorder: A systematic review and meta-analysis.Neuroscience. 2024 Sep 13;555:145-155. doi: 10.1016/j.neuroscience.2024.07.027. Epub 2024 Jul 25. Neuroscience. 2024. PMID: 39059741
Cited by
-
Canonical and non-canonical roles of oligodendrocyte precursor cells in mental disorders.Npj Ment Health Res. 2025 May 15;4(1):19. doi: 10.1038/s44184-025-00133-x. Npj Ment Health Res. 2025. PMID: 40374740 Free PMC article. Review.
-
Mitochondrial mass and low mitochondrial membrane potential percentage of CD8+ T cell subsets are implicated with therapeutic effect in depressive disorder.Front Psychiatry. 2025 Jul 18;16:1550958. doi: 10.3389/fpsyt.2025.1550958. eCollection 2025. Front Psychiatry. 2025. PMID: 40756310 Free PMC article.
References
-
- Zorrilla E. P., Luborsky L., McKay J. R., Rosenthal R., Houldin A., Tax A., McCorkle R., Seligman D. A., Schmidt K., Brain Behav. Immun. 2001, 15, 199. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
