How much metagenome data is needed for protein structure prediction: The advantages of targeted approach from the ecological and evolutionary perspectives
- PMID: 38867727
- PMCID: PMC10989767
- DOI: 10.1002/imt2.9
How much metagenome data is needed for protein structure prediction: The advantages of targeted approach from the ecological and evolutionary perspectives
Abstract
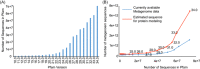
It has been proven that three-dimensional protein structures could be modeled by supplementing homologous sequences with metagenome sequences. Even though a large volume of metagenome data is utilized for such purposes, a significant proportion of proteins remain unsolved. In this review, we focus on identifying ecological and evolutionary patterns in metagenome data, decoding the complicated relationships of these patterns with protein structures, and investigating how these patterns can be effectively used to improve protein structure prediction. First, we proposed the metagenome utilization efficiency and marginal effect model to quantify the divergent distribution of homologous sequences for the protein family. Second, we proposed that the targeted approach effectively identifies homologous sequences from specified biomes compared with the untargeted approach's blind search. Finally, we determined the lower bound for metagenome data required for predicting all the protein structures in the Pfam database and showed that the present metagenome data is insufficient for this purpose. In summary, we discovered ecological and evolutionary patterns in the metagenome data that may be used to predict protein structures effectively. The targeted approach is promising in terms of effectively extracting homologous sequences and predicting protein structures using these patterns.
Keywords: ecology; evolution; metagenome data; protein 3D structure modeling; targeted approach.
© 2022 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Zhang, Chengxin , Zheng Wei, Freddolino Peter L., and Zhang Yang. 2018. “MetaGO: Predicting Gene Ontology of Non‐Homologous Proteins Through Low‐Resolution Protein Structure Prediction and Protein‐Protein Network Mapping.” Journal of Molecular Biology 430: 2256–65. 10.1016/j.jmb.2018.03.004 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
