hydro SIM: super-resolution speckle illumination microscopy with a hydrogel diffuser
- PMID: 38867780
- PMCID: PMC11166422
- DOI: 10.1364/BOE.521521
hydro SIM: super-resolution speckle illumination microscopy with a hydrogel diffuser
Abstract
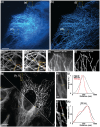
Super-resolution microscopy has emerged as an indispensable methodology for probing the intricacies of cellular biology. Structured illumination microscopy (SIM), in particular, offers an advantageous balance of spatial and temporal resolution, allowing for visualizing cellular processes with minimal disruption to biological specimens. However, the broader adoption of SIM remains hampered by the complexity of instrumentation and alignment. Here, we introduce speckle-illumination super-resolution microscopy using hydrogel diffusers (hydroSIM). The study utilizes the high scattering and optical transmissive properties of hydrogel materials and realizes a remarkably simplified approach to plug-in super-resolution imaging via a common epi-fluorescence platform. We demonstrate the hydroSIM system using various phantom and biological samples, and the results exhibited effective 3D resolution doubling, optical sectioning, and high contrast. We foresee hydroSIM, a cost-effective, biocompatible, and user-accessible super-resolution methodology, to significantly advance a wide range of biomedical imaging and applications.
© 2024 Optica Publishing Group.
Conflict of interest statement
The authors declare that there are no conflicts of interest in this article.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
