Molecular and clinical epidemiology of carbapenem resistant Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacterales in Fiji: a multicentre prospective observational study
- PMID: 38867891
- PMCID: PMC11166881
- DOI: 10.1016/j.lanwpc.2024.101095
Molecular and clinical epidemiology of carbapenem resistant Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacterales in Fiji: a multicentre prospective observational study
Abstract
Background: Carbapenem resistant organisms (CROs) such as Acinetobacter baumannii (CRAb), Pseudomonas aeruginosa (CRPa), Escherichia coli (CREc), and Klebsiella pneumoniae (CRKp) have been identified by the World Health Organization (WHO) as global priority pathogens. The dissemination of these pathogens and clonal outbreaks within healthcare facilities are of serious concern, particularly in regions with limited resources. In Fiji, where healthcare services are primarily provided by public hospitals, understanding the extent and nature of this problem is essential for the development of effective patient management, prevention interventions and control strategies.
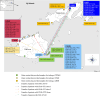
Methods: CROs isolated from 211 (77.3%) non-sterile (urinary catheters, urine, sputum, wound swab, and endotracheal tube) and 62 (22.7%) normally sterile (blood, cerebrospinal fluid, intravascular catheter, and aspirates) body sites of 272 patients treated at the three major hospitals in Fiji, the Colonial War Memorial Hospital (CWMH), Lautoka Hospital (LTKH), and Labasa Hospital (LBSH), and outer peripheral health centres around Fiji, were analysed. Clinical and demographic patient data such as age, sex, admission diagnosis, admission and discharge dates, patient outcomes, date of death, start and end date of meropenem and colistin treatment were reviewed. These CRO isolates comprised A. baumannii, P. aeruginosa, E. coli, and K. pneumoniae, that were prospectively collected at the microbiology laboratory of CWMH and LBSH from January 2020 through August 2021 and at the LTKH from January 2020 to December 2021. In addition, 10 retrospectively stored CRPa isolates collected from patients at the CWMH from January through December 2019, were also included in the study. All isolates were characterised using mass spectrometry, antimicrobial susceptibility testing, and whole genome sequencing. Phylogenetic relationships among the CROs were assessed through core genome single nucleotide polymorphism (SNP) analysis. The CRAb isolates were also compared to the CRAb isolates from CWMH isolated in 2016/2017 and 2019, along with CRAb isolates obtained from Fijian patients admitted to New Zealand hospitals in 2020 and 2021 from our retrospective study.
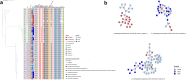
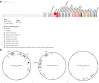
Findings: Of 272 patients, 140 (51.5%) were male, the median (range) age of patients was 45 (<1-89) years, 161 (59.2%) were I-Taukei, 104 (38.2%) Fijians of Indian descent, and 7 (2.6%) were from other ethnic backgrounds. 234 (86.0%) of these 272 patients, had their first positive CRO sample collected ≥72 h following admission and the remaining 38 (14.0%) were isolated within 72 h following admission. Of the 273 CROs, 146 (53.5%) were collected at the CWMH, 66 (24.2%) LTKH, and 61 (22.3%) LBSH, while 62 (22.7%) were isolated from normally sterile sites and 211 (77.3%) from sites that are not sterile. Of 273 isolates, 131 (48.0%) were CRAb, 90 (33.0%) CRPa, 46 (16.8%) CREc, and 6 (2.2%) CRKp. Of 131 CRAb, 108 (82.4%) were ST2, with three distinct clones, all encoding bla OXA-23 and bla OXA - 66, while clone 3 also encoded bla NDM-1; bla OXA-23 was associated with two copies of ISAba1 insertion element, forming the composite transposon Tn2006. The first two CRAb ST2 clones were genetically linked to those isolated at CMWH 2016 through 2019, while the third was genetically linked to isolates from Fijian patients admitted to New Zealand hospitals in 2020 and 2021. Of CRPa, 65 (72.2%) were ST773 and carried β-lactamase genes bla NDM-1, bla OXA-50, and bla OXA-395. Of 10 retrospective CRPa isolates, all belonged to CRPa ST773 and carried bla NDM-1, bla OXA-50, and bla OXA-395. Of 46 CREc, 44 (95.7%) were ST410 and encoded bla NDM-7 on an IncX3 plasmid. Of 6 CRKp, 4 (66.7%) were ST16 and carried bla NDM-5 on an IncX3 plasmid. Other sequence types of CRPa (ST9, ST357, ST654, ST664), CRAb (ST25, ST374, ST499), CREc (ST167), and CRKp (ST45, ST336) were also detected. Of those receiving meropenem treatment in the prospective study, 30 (57.7%) received it inappropriately. Of 272 patients, 65 (23.9%) died within the 30 days after first positive CRO isolation.
Interpretation: We identified nosocomial transmission of distinct clones of CRAb ST2, CRPa ST773, CREc ST410, and CRKp ST16 within and between the three major hospitals in Fiji. Moreover, community onset infections associated with CRPa, CREc, and CRAb were also detected. Of note, cross-border transmission of CRAb ST2 clone 3 strain between Fiji and New Zealand was also detected. These clones encoded an array of carbapenem resistance genes associated with mobile genetic elements, including plasmids, transposons, and integrative and conjugative elements, signifying their potential for increased mobility, further acquisition of resistance genes, and spread. Inappropriate use of meropenem was common. Of note, the majority of patients who died had acquired CRO during their hospital stay. These findings highlight the need for stringent IPC strategies focusing on catheter and ventilator management, meticulous wound care, rigorous sepsis control, consistent hand hygiene, effective use of disinfectants, and thorough sanitisation of both hospital environments and medical equipment in the three major hospitals in Fiji. Additionally, diligent surveillance of AMR and robust antimicrobial stewardship are crucial for effectively managing nosocomial infections.
Funding: This project was funded by the Otago Medical School Foundations Trust (Dean's Bequest Fund) and a Fiji National University seed grant. The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Keywords: Acinetobacter baumannii; Carbapenem resistant; Escherichia coli; Klebsiella pneumoniae; Pseudomonas aeruginosa.
© 2024 The Author(s).
Conflict of interest statement
The authors have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Environmental contamination with carbapenem resistant Acinetobacter baumannii in healthcare settings in Fiji: a potential source of infection.Front Cell Infect Microbiol. 2024 Sep 23;14:1429443. doi: 10.3389/fcimb.2024.1429443. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39376664 Free PMC article.
-
Molecular and clinical epidemiology of carbapenem resistant Acinetobacter baumannii ST2 in Oceania: a multicountry cohort study.Lancet Reg Health West Pac. 2023 Sep 22;40:100896. doi: 10.1016/j.lanwpc.2023.100896. eCollection 2023 Nov. Lancet Reg Health West Pac. 2023. PMID: 38116498 Free PMC article.
-
World Health Organization priority antimicrobial resistance in Enterobacterales, Acinetobacter baumannii, Pseudomonas aeruginosa, Staphylococcus aureus and Enterococcus faecium healthcare-associated bloodstream infections in Brazil (ASCENSION): a prospective, multicentre, observational study.Lancet Reg Health Am. 2025 Jan 30;43:101004. doi: 10.1016/j.lana.2025.101004. eCollection 2025 Mar. Lancet Reg Health Am. 2025. PMID: 39957800 Free PMC article.
-
Hospital and municipal wastewater as a source of carbapenem-resistant Acinetobacter baumannii and Pseudomonas aeruginosa in the environment: a review.Environ Sci Pollut Res Int. 2024 Aug;31(36):48813-48838. doi: 10.1007/s11356-024-34436-x. Epub 2024 Jul 25. Environ Sci Pollut Res Int. 2024. PMID: 39052110 Free PMC article. Review.
-
Mortality and genetic diversity of antibiotic-resistant bacteria associated with bloodstream infections: a systemic review and genomic analysis.BMC Infect Dis. 2024 Dec 4;24(1):1385. doi: 10.1186/s12879-024-10274-7. BMC Infect Dis. 2024. PMID: 39633294 Free PMC article.
Cited by
-
Genomic diversity of clinically relevant bacterial pathogens from an acute care hospital in Suva, Fiji.JAC Antimicrob Resist. 2025 Jun 9;7(3):dlaf058. doi: 10.1093/jacamr/dlaf058. eCollection 2025 Jun. JAC Antimicrob Resist. 2025. PMID: 40492256 Free PMC article.
-
Establishing carbapenem resistant organism surveillance, prevention, and control in a middle-income country: implementation of a hospital-based program in Fiji.Antimicrob Resist Infect Control. 2025 Mar 5;14(1):19. doi: 10.1186/s13756-025-01534-5. Antimicrob Resist Infect Control. 2025. PMID: 40038841 Free PMC article.
-
Environmental contamination with carbapenem resistant Acinetobacter baumannii in healthcare settings in Fiji: a potential source of infection.Front Cell Infect Microbiol. 2024 Sep 23;14:1429443. doi: 10.3389/fcimb.2024.1429443. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39376664 Free PMC article.
-
Diverse modes of ceftazidime/avibactam resistance acquisition in carbapenem-resistant Klebsiella pneumoniae and Pseudomonas aeruginosa from a Chinese intensive care unit.Ann Clin Microbiol Antimicrob. 2025 May 30;24(1):35. doi: 10.1186/s12941-025-00800-z. Ann Clin Microbiol Antimicrob. 2025. PMID: 40448249 Free PMC article.
-
Complex Infection-Control Measures with Disinfectant Switch Help the Successful Early Control of Carbapenem-Resistant Acinetobacter baumannii Outbreak in Intensive Care Unit.Antibiotics (Basel). 2024 Sep 11;13(9):869. doi: 10.3390/antibiotics13090869. Antibiotics (Basel). 2024. PMID: 39335042 Free PMC article.
References
-
- Pan Y.-P., Xu Y.-H., Wang Z.-X., Fang Y.-P., Shen J.-L. Overexpression of MexAB-OprM efflux pump in carbapenem-resistant Pseudomonas aeruginosa. Arch Microbiol. 2016;198(6):565–571. - PubMed
LinkOut - more resources
Full Text Sources

