Metabolite profiling of human-originated Lachnospiraceae at the strain level
- PMID: 38867908
- PMCID: PMC10989990
- DOI: 10.1002/imt2.58
Metabolite profiling of human-originated Lachnospiraceae at the strain level
Abstract
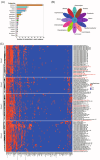
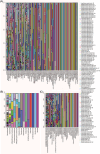
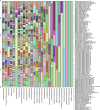
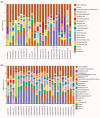
The human gastrointestinal (GI) tract harbors diverse microbes, and the family Lachnospiraceae is one of the most abundant and widely occurring bacterial groups in the human GI tract. Beneficial and adverse effects of the Lachnospiraceae on host health were reported, but the diversities at species/strain levels as well as their metabolites of Lachnospiraceae have been, so far, not well documented. In the present study, we report on the collection of 77 human-originated Lachnospiraceae species (please refer hLchsp, https://hgmb.nmdc.cn/subject/lachnospiraceae) and the in vitro metabolite profiles of 110 Lachnospiraceae strains (https://hgmb.nmdc.cn/subject/lachnospiraceae/metabolites). The Lachnospiraceae strains in hLchsp produced 242 metabolites of 17 categories. The larger categories were alcohols (89), ketones (35), pyrazines (29), short (C2-C5), and long (C > 5) chain acids (31), phenols (14), aldehydes (14), and other 30 compounds. Among them, 22 metabolites were aromatic compounds. The well-known beneficial gut microbial metabolite, butyric acid, was generally produced by many Lachnospiraceae strains, and Agathobacter rectalis strain Lach-101 and Coprococcus comes strain NSJ-173 were the top 2 butyric acid producers, as 331.5 and 310.9 mg/L of butyric acids were produced in vitro, respectively. Further analysis of the publicly available cohort-based volatile-metabolomic data sets of human feces revealed that over 30% of the prevailing volatile metabolites were covered by Lachnospiraceae metabolites identified in this study. This study provides Lachnospiraceae strain resources together with their metabolic profiles for future studies on host-microbe interactions and developments of novel probiotics or biotherapies.
Keywords: Blautia; Lachnospiraceae; alcohols; aldehydes; metabolite profiling; phenols; short‐chain fatty acids.
© 2022 The Authors. iMeta published by John Wiley & Sons Australia Ltd. on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Dietary glycation compounds - implications for human health.Crit Rev Toxicol. 2024 Sep;54(8):485-617. doi: 10.1080/10408444.2024.2362985. Epub 2024 Aug 16. Crit Rev Toxicol. 2024. PMID: 39150724
-
Enlightening the taxonomy darkness of human gut microbiomes with a cultured biobank.Microbiome. 2021 May 21;9(1):119. doi: 10.1186/s40168-021-01064-3. Microbiome. 2021. PMID: 34020714 Free PMC article.
-
Pararoseburia lenta gen. nov., sp. nov. isolated from human faeces.Int J Syst Evol Microbiol. 2022 May;72(5). doi: 10.1099/ijsem.0.005371. Int J Syst Evol Microbiol. 2022. PMID: 35559790
-
Microbiome-Metabolomics Analysis of the Impacts of Cryptosporidium muris Infection in BALB/C Mice.Microbiol Spectr. 2023 Feb 14;11(1):e0217522. doi: 10.1128/spectrum.02175-22. Epub 2022 Dec 19. Microbiol Spectr. 2023. PMID: 36533947 Free PMC article.
-
The Controversial Role of Human Gut Lachnospiraceae.Microorganisms. 2020 Apr 15;8(4):573. doi: 10.3390/microorganisms8040573. Microorganisms. 2020. PMID: 32326636 Free PMC article. Review.
Cited by
-
Repressed Blautia-acetate immunological axis underlies breast cancer progression promoted by chronic stress.Nat Commun. 2023 Oct 3;14(1):6160. doi: 10.1038/s41467-023-41817-2. Nat Commun. 2023. PMID: 37789028 Free PMC article.
-
Insight into the Potential of Somatostatin Vaccination with Goats as a Model: From a Perspective of the Gastrointestinal Microbiota.Animals (Basel). 2025 Mar 4;15(5):728. doi: 10.3390/ani15050728. Animals (Basel). 2025. PMID: 40076011 Free PMC article.
-
Enhanced propionate and butyrate metabolism in cecal microbiota contributes to cold-stress adaptation in sheep.Microbiome. 2025 Apr 24;13(1):103. doi: 10.1186/s40168-025-02096-9. Microbiome. 2025. PMID: 40275300 Free PMC article.
-
mKmer: an unbiased K-mer embedding of microbiomic single-microbe RNA sequencing data.Brief Bioinform. 2025 May 1;26(3):bbaf227. doi: 10.1093/bib/bbaf227. Brief Bioinform. 2025. PMID: 40407385 Free PMC article.
-
Gut Microbiota-Butyrate-PPARγ Axis Modulates Adipose Regulatory T Cell Population.Adv Sci (Weinh). 2025 May;12(20):e2411086. doi: 10.1002/advs.202411086. Epub 2025 Feb 25. Adv Sci (Weinh). 2025. PMID: 39998325 Free PMC article.
References
-
- Almeida, Alexandre , Nayfach Stephen, Boland Miguel, Strozzi Francesco, Beracochea Martin, Shi Zhou Jason, Pollard Katherine S., et al. 2021. “A Unified Catalog of 204,938 Reference Genomes from the Human Gut Microbiome.” Nature Biotechnology 39: 105–14. 10.1038/s41587-020-0603-3 - DOI - PMC - PubMed
-
- Tyakht, Alexander V. , Kostryukova Elena S., Popenko Anna S., Belenikin Maxim S., Pavlenko Alexander V., Larin Andrey K., Karpova Irina Y., et al. 2013. “Human Gut Microbiota Community Structures in Urban and Rural Populations in Russia.” Nature Communications 4: 2469. 10.1038/ncomms3469 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
