Gut microbiota composition in the sympatric and diet-sharing Drosophila simulans and Dicranocephalus wallichii bowringi shaped largely by community assembly processes rather than regional species pool
- PMID: 38867909
- PMCID: PMC10989964
- DOI: 10.1002/imt2.57
Gut microbiota composition in the sympatric and diet-sharing Drosophila simulans and Dicranocephalus wallichii bowringi shaped largely by community assembly processes rather than regional species pool
Abstract
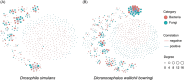
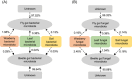
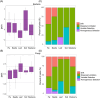
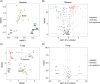
Clarifying the mechanisms underlying microbial community assembly from regional microbial pools is a central issue of microbial ecology, but remains largely unexplored. Here, we investigated the gut bacterial and fungal microbiome assembly processes and potential sources in Drosophila simulans and Dicranocephalus wallichii bowringi, two wild, sympatric insect species that share a common diet of waxberry. While some convergence was observed, the diversity, composition, and network structure of the gut microbiota significantly differed between these two host species. Null model analyses revealed that stochastic processes (e.g., drift, dispersal limitation) play a principal role in determining gut microbiota from both hosts. However, the strength of each ecological process varied with the host species. Furthermore, the source-tracking analysis showed that only a minority of gut microbiota within D. simulans and D. wallichii bowringi are drawn from a regional microbial pool from waxberries, leaves, or soil. Results from function prediction implied that host species-specific gut microbiota might arise partly through host functional requirement and specific selection across host-microbiota coevolution. In conclusion, our findings uncover the importance of community assembly processes over regional microbial pools in shaping sympatric insect gut microbiome structure and function.
Keywords: Dicranocephalus wallichii bowringi; Drosophila simulans; community assembly; gut microbiota; microbial source‐tracking.
© 2022 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Stochastic processes drive divergence of bacterial and fungal communities in sympatric wild insect species despite sharing a common diet.mSphere. 2024 Aug 28;9(8):e0038624. doi: 10.1128/msphere.00386-24. Epub 2024 Aug 6. mSphere. 2024. PMID: 39105581 Free PMC article.
-
Species Identity Dominates over Environment in Driving Bacterial Community Assembly in Wild Invasive Leaf Miners.Microbiol Spectr. 2022 Apr 27;10(2):e0026622. doi: 10.1128/spectrum.00266-22. Epub 2022 Mar 28. Microbiol Spectr. 2022. PMID: 35343791 Free PMC article.
-
Host Species and Geography Differentiate Honeybee Gut Bacterial Communities by Changing the Relative Contribution of Community Assembly Processes.mBio. 2021 Jun 29;12(3):e0075121. doi: 10.1128/mBio.00751-21. Epub 2021 Jun 1. mBio. 2021. PMID: 34061602 Free PMC article.
-
Integrating species traits into species pools.Ecology. 2018 Jun;99(6):1265-1276. doi: 10.1002/ecy.2220. Ecology. 2018. PMID: 29569239 Review.
-
Global landscape of gut microbiome diversity and antibiotic resistomes across vertebrates.Sci Total Environ. 2022 Sep 10;838(Pt 2):156178. doi: 10.1016/j.scitotenv.2022.156178. Epub 2022 May 23. Sci Total Environ. 2022. PMID: 35618126 Review.
Cited by
-
The giant panda gut harbors a high diversity of lactic acid bacteria revealed by a novel culturomics pipeline.mSystems. 2024 Jul 23;9(7):e0052024. doi: 10.1128/msystems.00520-24. Epub 2024 Jun 26. mSystems. 2024. PMID: 38920380 Free PMC article.
-
Computer-aided analysis reveals metallothionein-positive cancer-associated fibroblasts promote angiogenesis in gastric adenocarcinoma.Discov Oncol. 2024 Dec 5;15(1):751. doi: 10.1007/s12672-024-01614-9. Discov Oncol. 2024. PMID: 39636347 Free PMC article.
-
ZC3H12D gene expression exhibits dual effects on the development and progression of lung adenocarcinoma.Sci Rep. 2025 May 18;15(1):17234. doi: 10.1038/s41598-025-02163-z. Sci Rep. 2025. PMID: 40383849 Free PMC article.
-
CTSG is a prognostic marker involved in immune infiltration and inhibits tumor progression though the MAPK signaling pathway in non-small cell lung cancer.J Cancer Res Clin Oncol. 2024 Dec 26;151(1):21. doi: 10.1007/s00432-024-06051-3. J Cancer Res Clin Oncol. 2024. PMID: 39724501 Free PMC article.
-
Dynamic response of gut microbiota mediates the adaptation of Cipangopaludina chinensis to Pomacea canaliculata invasion.Microbiome. 2025 Jul 24;13(1):171. doi: 10.1186/s40168-025-02160-4. Microbiome. 2025. PMID: 40708023 Free PMC article.
References
-
- Gould, Alison L. , Zhang Vivian, Lamberti Lisa, Jones Eric W., Obadia Benjamin, Korasidis Nikolaos, and Gavryushkin Alex.2018. “Microbiome Interactions Shape Host Fitness.” Proceedings of the National Academy of Sciences of the United States of America 115: 11951–60. 10.1073/pnas.1809349115 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
