Cultivation strategies for prokaryotes from extreme environments
- PMID: 38867929
- PMCID: PMC10989778
- DOI: 10.1002/imt2.123
Cultivation strategies for prokaryotes from extreme environments
Abstract
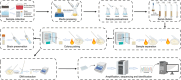
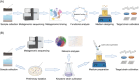
The great majority of microorganisms are as-yet-uncultivated, mostly found in extreme environments. High-throughput sequencing provides data-rich genomes from single-cell and metagenomic techniques, which has enabled researchers to obtain a glimpse of the unexpected genetic diversity of "microbial dark matter." However, cultivating microorganisms from extreme environments remains essential for dissecting and utilizing the functions of extremophiles. Here, we provide a straightforward protocol for efficiently isolating prokaryotic microorganisms from different extreme habitats (thermal, xeric, saline, alkaline, acidic, and cryogenic environments), which was established through previous successful work and our long-term experience in extremophile resource mining. We propose common processes for extremophile isolation at first and then summarize multiple cultivation strategies for recovering prokaryotic microorganisms from extreme environments and meanwhile provide specific isolation tips that are always overlooked but important. Furthermore, we propose the use of multi-omics-guided microbial cultivation approaches for culturing these as-yet-uncultivated microorganisms and two examples are provided to introduce how these approaches work. In summary, the protocol allows researchers to significantly improve the isolation efficiency of pure cultures and novel taxa, which therefore paves the way for the protection and utilization of microbial resources from extreme environments.
Keywords: cultivation strategy; extreme environment; extremophiles; multi‐omics; pure culture.
© 2023 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Mining microbial and metabolic dark matter in extreme environments: a roadmap for harnessing the power of multi-omics data.Adv Biotechnol (Singap). 2024 Aug 5;2(3):26. doi: 10.1007/s44307-024-00034-8. Adv Biotechnol (Singap). 2024. PMID: 39883228 Free PMC article. Review.
-
Shedding light on the composition of extreme microbial dark matter: alternative approaches for culturing extremophiles.Front Microbiol. 2023 Jun 2;14:1167718. doi: 10.3389/fmicb.2023.1167718. eCollection 2023. Front Microbiol. 2023. PMID: 37333658 Free PMC article. Review.
-
A comprehensive review on chromium (Cr) contamination and Cr(VI)-resistant extremophiles in diverse extreme environments.Environ Sci Pollut Res Int. 2023 May;30(21):59163-59193. doi: 10.1007/s11356-023-26624-y. Epub 2023 Apr 13. Environ Sci Pollut Res Int. 2023. PMID: 37046169 Review.
-
Bioprospecting of Novel Extremozymes From Prokaryotes-The Advent of Culture-Independent Methods.Front Microbiol. 2021 Feb 10;12:630013. doi: 10.3389/fmicb.2021.630013. eCollection 2021. Front Microbiol. 2021. PMID: 33643258 Free PMC article. Review.
-
The multi-omics promise in context: from sequence to microbial isolate.Crit Rev Microbiol. 2018 Mar;44(2):212-229. doi: 10.1080/1040841X.2017.1332003. Epub 2017 May 31. Crit Rev Microbiol. 2018. PMID: 28562180 Review.
Cited by
-
Cultivation of novel Atribacterota from oil well provides new insight into their diversity, ecology, and evolution in anoxic, carbon-rich environments.Microbiome. 2024 Jul 6;12(1):123. doi: 10.1186/s40168-024-01836-7. Microbiome. 2024. PMID: 38971798 Free PMC article.
-
Mining microbial and metabolic dark matter in extreme environments: a roadmap for harnessing the power of multi-omics data.Adv Biotechnol (Singap). 2024 Aug 5;2(3):26. doi: 10.1007/s44307-024-00034-8. Adv Biotechnol (Singap). 2024. PMID: 39883228 Free PMC article. Review.
-
The family Thermoactinomycetaceae: an emerging microbial resource with high application value.Front Microbiol. 2025 Jan 28;16:1507902. doi: 10.3389/fmicb.2025.1507902. eCollection 2025. Front Microbiol. 2025. PMID: 39935634 Free PMC article. Review.
-
Hohaiivirga grylli gen. nov., sp. nov., a New Member of the Family Methylobacteriaceae, Isolated from Cricket (Gryllus chinensis).Curr Microbiol. 2024 Oct 6;81(11):392. doi: 10.1007/s00284-024-03922-3. Curr Microbiol. 2024. PMID: 39369359
-
Roseobacter sinensis sp. nov., a marine bacterium capable to synthesize arachidonic acid.Antonie Van Leeuwenhoek. 2024 Oct 29;118(1):24. doi: 10.1007/s10482-024-02034-z. Antonie Van Leeuwenhoek. 2024. PMID: 39472387
References
LinkOut - more resources
Full Text Sources
