Single-nucleus transcriptomics reveal cardiac cell type-specific diversification in metabolic disease transgenic pigs
- PMID: 38868189
- PMCID: PMC11166884
- DOI: 10.1016/j.isci.2024.110015
Single-nucleus transcriptomics reveal cardiac cell type-specific diversification in metabolic disease transgenic pigs
Abstract
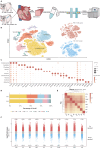
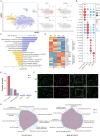
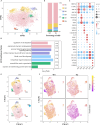
Cardiac damage is widely present in patients with metabolic diseases, but the exact pathophysiological mechanisms involved remain unclear. The porcine heart is an ideal material for cardiovascular research due to its similarities to the human heart. This study evaluated pathological features and performed single-nucleus RNA sequencing (snRNA-seq) on myocardial samples from both wild-type and metabolic disease-susceptible transgenic pigs (previously established). We found that transgenic pigs exhibited lipid metabolism disturbances and myocardial injury after a high-fat high-sucrose diet intervention. snRNA-seq reveals the cellular landscape of healthy and metabolically disturbed pig hearts and identifies the major cardiac cell populations affected by metabolic diseases. Within metabolic disorder hearts, metabolically active cardiomyocytes exhibited impaired function and reduced abundance. Moreover, massive numbers of reparative LYVE1+ macrophages were lost. Additionally, proinflammatory endothelial cells were activated with high expression of multiple proinflammatory cytokines. Our findings provide insights into the cellular mechanisms of metabolic disease-induced myocardial injury.
Keywords: Animals; Integrative aspects of cell biology; Model organism; Porcine cardiology; Transcriptomics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
SnRNA-Seq of Pancreas Revealed the Dysfunction of Endocrine and Exocrine Cells in Transgenic Pigs with Prediabetes.Int J Mol Sci. 2023 Apr 22;24(9):7701. doi: 10.3390/ijms24097701. Int J Mol Sci. 2023. PMID: 37175407 Free PMC article.
-
Single-nucleus transcriptomic survey of cell diversity and functional maturation in postnatal mammalian hearts.Genes Dev. 2018 Oct 1;32(19-20):1344-1357. doi: 10.1101/gad.316802.118. Epub 2018 Sep 25. Genes Dev. 2018. PMID: 30254108 Free PMC article.
-
Transgenic expression of the human A20 gene in cloned pigs provides protection against apoptotic and inflammatory stimuli.Xenotransplantation. 2009 Nov-Dec;16(6):522-34. doi: 10.1111/j.1399-3089.2009.00556.x. Xenotransplantation. 2009. PMID: 20042052
-
Cooperation of liver cells in health and disease.Adv Anat Embryol Cell Biol. 2001;161:III-XIII, 1-151. doi: 10.1007/978-3-642-56553-3. Adv Anat Embryol Cell Biol. 2001. PMID: 11729749 Review.
-
Recent advances in deciphering hippocampus complexity using single-cell transcriptomics.Neurobiol Dis. 2023 Apr;179:106062. doi: 10.1016/j.nbd.2023.106062. Epub 2023 Mar 4. Neurobiol Dis. 2023. PMID: 36878328 Review.
Cited by
-
Transcriptomic signatures of atheroresistance in the human atrium and ventricle highlight potential candidates for targeted atherosclerosis therapeutics.Biochem Biophys Rep. 2025 Apr 8;42:102007. doi: 10.1016/j.bbrep.2025.102007. eCollection 2025 Jun. Biochem Biophys Rep. 2025. PMID: 40248137 Free PMC article.
References
-
- McLellan M.A., Skelly D.A., Dona M.S.I., Squiers G.T., Farrugia G.E., Gaynor T.L., Cohen C.D., Pandey R., Diep H., Vinh A., et al. High-Resolution Transcriptomic Profiling of the Heart During Chronic Stress Reveals Cellular Drivers of Cardiac Fibrosis and Hypertrophy. Circulation. 2020;142:1448–1463. doi: 10.1161/CIRCULATIONAHA.119.045115. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

