Lack of memory recall in human CD4 T cells elicited by the first encounter with SARS-CoV-2
- PMID: 38868209
- PMCID: PMC11166706
- DOI: 10.1016/j.isci.2024.109992
Lack of memory recall in human CD4 T cells elicited by the first encounter with SARS-CoV-2
Abstract
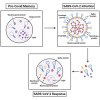
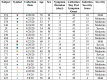
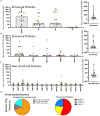
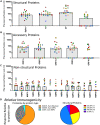
The studies reported here focus on the impact of pre-existing CD4 T cell immunity on the first encounter with SARS-CoV-2. They leverage PBMC samples from plasma donors collected after a first SARS-CoV-2 infection, prior to vaccine availability and compared to samples collected prior to the emergence of SARS-CoV-2. Analysis of CD4 T cell specificity across the entire SARS-CoV-2 proteome revealed that the recognition of SARS-CoV-2-derived epitopes by CD4 memory cells prior to the pandemic are enriched for reactivity toward non-structural proteins conserved across endemic CoV strains. However, CD4 T cells after primary infection with SARS-CoV-2 focus on epitopes from structural proteins. We observed little evidence for preferential recall to epitopes conserved between SARS-CoV-2 and seasonal CoV, a finding confirmed through use of selectively curated conserved and SARS-unique peptides. Our data suggest that SARS-CoV-2 CD4 T cells elicited by the first infection are primarily established from the naive CD4 T cell pool.
Keywords: Health sciences; Immune response; Immunology; Virology.
© 2024 The Author(s).
Conflict of interest statement
P.G.T. has consulted or received travel support from JNJ, Pfizer, Illumina, 10X Genomics, Merck, and PACT Pharma and served on the SAB of Immunoscape, Shennon Bio, and Cytoagents. No other authors have competing interests.
Figures

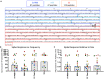

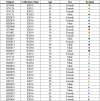
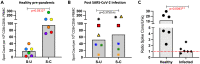

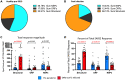
References
-
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. - DOI - PMC - PubMed
-
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100204. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

