MetaSVs: A pipeline combining long and short reads for analysis and visualization of structural variants in metagenomes
- PMID: 38868213
- PMCID: PMC10989790
- DOI: 10.1002/imt2.139
MetaSVs: A pipeline combining long and short reads for analysis and visualization of structural variants in metagenomes
Abstract
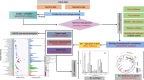
Structural variants (SVs, including large-scale insertions, deletions, inversions, and translocations) significantly impact the functions of genes in the microbial genome, and SVs in the microbiome are associated with diverse biological processes and human diseases. With the advancements in sequencing and bioinformatics technologies, increasingly, sequencing data and analysis tools are already being extensively utilized for microbiome SV analyses, leading to a higher demand for more dedicated SV analysis workflows. Moreover, due to the unique detection biases of various sequencing technologies, including short-read sequencing (such as Illumina platforms) and long-read sequencing (e.g., Oxford Nanopore and PacBio), SV discovery based on multiple platforms is necessary to comprehensively identify the wide variety of SVs. Here, we establish an integrated pipeline MetaSVs combining Nanopore long reads and Illumina short reads to analyze SVs in the microbial genomes from gut microbiome and further identify differential SVs that can be reflective of metabolic differences. Our pipeline provides researchers easy access to SVs and relevant metabolites in the microbial genomes without the requirement of specific technical expertise, which is particularly useful to researchers interested in metagenomic SVs but lacking sophisticated bioinformatic knowledge.
Keywords: hybrid sequencing; metagenome; microbiome; nanopore; structural variants.
© 2023 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Combined use of Oxford Nanopore and Illumina sequencing yields insights into soybean structural variation biology.BMC Biol. 2022 Feb 23;20(1):53. doi: 10.1186/s12915-022-01255-w. BMC Biol. 2022. PMID: 35197050 Free PMC article.
-
Comparison of structural variants detected by PacBio-CLR and ONT sequencing in pear.BMC Genomics. 2022 Dec 14;23(1):830. doi: 10.1186/s12864-022-09074-7. BMC Genomics. 2022. PMID: 36517766 Free PMC article.
-
Benchmarking Oxford Nanopore read alignment-based insertion and deletion detection in crop plant genomes.Plant Genome. 2023 Jun;16(2):e20314. doi: 10.1002/tpg2.20314. Epub 2023 Mar 29. Plant Genome. 2023. PMID: 36988043
-
Resolving complex structural variants via nanopore sequencing.Front Genet. 2023 Aug 16;14:1213917. doi: 10.3389/fgene.2023.1213917. eCollection 2023. Front Genet. 2023. PMID: 37674481 Free PMC article. Review.
-
Application of long-read sequencing to the detection of structural variants in human cancer genomes.Comput Struct Biotechnol J. 2021 Jul 28;19:4207-4216. doi: 10.1016/j.csbj.2021.07.030. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34527193 Free PMC article. Review.
Cited by
-
Reference-free structural variant detection in microbiomes via long-read co-assembly graphs.Bioinformatics. 2024 Jun 28;40(Suppl 1):i58-i67. doi: 10.1093/bioinformatics/btae224. Bioinformatics. 2024. PMID: 38940156 Free PMC article.
-
Pangenome and genome variation analyses of pigs unveil genomic facets for their adaptation and agronomic characteristics.Imeta. 2024 Dec 26;3(6):e257. doi: 10.1002/imt2.257. eCollection 2024 Dec. Imeta. 2024. PMID: 39742300 Free PMC article.
-
Reference-free Structural Variant Detection in Microbiomes via Long-read Coassembly Graphs.bioRxiv [Preprint]. 2024 Jan 30:2024.01.25.577285. doi: 10.1101/2024.01.25.577285. bioRxiv. 2024. Update in: Bioinformatics. 2024 Jun 28;40(Suppl 1):i58-i67. doi: 10.1093/bioinformatics/btae224. PMID: 38352454 Free PMC article. Updated. Preprint.
References
-
- Yassour, Moran , Vatanen Tommi, Siljander Heli, Hämäläinen Anu‐Maaria, Härkönen Taina, Ryhänen Samppa J., Franzosa Eric A., et al. 2016. “Natural History of the Infant Gut Microbiome and Impact of Antibiotic Treatment on Bacterial Strain Diversity and Stability.” Science Translational Medicine 8: 343ra381. 10.1126/scitranslmed.aad0917 - DOI - PMC - PubMed
-
- Pärnänen, Katariina , Karkman Antti, Hultman Jenni, Lyra Christina, Bengtsson‐Palme Johan, Larsson D. G. Joakim, Rautava Samuli, et al. 2018. “Maternal Gut and Breast Milk Microbiota Affect Infant Gut Antibiotic Resistome and Mobile Genetic Elements.” Nature Communications 9: 3891. 10.1038/s41467-018-06393-w - DOI - PMC - PubMed
-
- Leung, Marcus H. Y. , Tong Xinzhao, Bastien Philippe, Guinot Florent, Tenenhaus Arthur, Appenzeller Brice M. R., Betts Richard J., et al. 2020. “Changes of the Human Skin Microbiota Upon Chronic Exposure to Polycyclic Aromatic Hydrocarbon Pollutants.” Microbiome 8: 100. 10.1186/s40168-020-00874-1 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
