VeloPro: A pipeline integrating Ribo-seq and AlphaFold deciphers association patterns between translation velocity and protein structure features
- PMID: 38868219
- PMCID: PMC10989810
- DOI: 10.1002/imt2.148
VeloPro: A pipeline integrating Ribo-seq and AlphaFold deciphers association patterns between translation velocity and protein structure features
Abstract
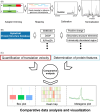
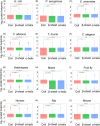
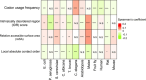
VeloPro integrates Ribo-seq data and AlphaFold2-predicted 3D protein structure information for characterization of the association patterns between translation velocity and many protein structure features in prokaryotic and eukaryotic organisms across different taxonomical clades such as bacteria, fungi, protozoa, nematode, plants, insect, and mammals. We illustrated that association patterns between translation velocity and protein structure features differ across organisms, partially reflecting their taxonomical relationship.
© 2023 The Authors. iMeta published by John Wiley & Sons Australia, Ltd on behalf of iMeta Science.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
GWIPS-viz: development of a ribo-seq genome browser.Nucleic Acids Res. 2014 Jan;42(Database issue):D859-64. doi: 10.1093/nar/gkt1035. Epub 2013 Oct 31. Nucleic Acids Res. 2014. PMID: 24185699 Free PMC article.
-
Structured RNA Contaminants in Bacterial Ribo-Seq.mSphere. 2020 Oct 21;5(5):e00855-20. doi: 10.1128/mSphere.00855-20. mSphere. 2020. PMID: 33087519 Free PMC article.
-
The ribosome profiling landscape of yeast reveals a high diversity in pervasive translation.Genome Biol. 2024 Oct 14;25(1):268. doi: 10.1186/s13059-024-03403-7. Genome Biol. 2024. PMID: 39402662 Free PMC article.
-
A review of Ribosome profiling and tools used in Ribo-seq data analysis.Comput Struct Biotechnol J. 2024 Apr 22;23:1912-1918. doi: 10.1016/j.csbj.2024.04.051. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38721586 Free PMC article. Review.
-
Beyond Read-Counts: Ribo-seq Data Analysis to Understand the Functions of the Transcriptome.Trends Genet. 2017 Oct;33(10):728-744. doi: 10.1016/j.tig.2017.08.003. Epub 2017 Sep 5. Trends Genet. 2017. PMID: 28887026 Review.
Cited by
-
Visualizing set relationships: EVenn's comprehensive approach to Venn diagrams.Imeta. 2024 Apr 11;3(3):e184. doi: 10.1002/imt2.184. eCollection 2024 Jun. Imeta. 2024. PMID: 38898979 Free PMC article.
References
-
- Samatova, Ekaterina , Daberger Jan, Liutkute Marija, and Rodnina Marina V.. 2021. “Translational Control by Ribosome Pausing in Bacteria: How a Non‐uniform Pace of Translation Affects Protein Production and Folding.” Frontiers in Microbiology 11: 619430. 10.3389/fmicb.2020.619430 - DOI - PMC - PubMed
-
- Buhr, Florian , Jha Sujata, Thommen Michael, Mittelstaet Joerg, Kutz Felicitas, Schwalbe Harald, Rodnina Marina V., and Komar Anton A.. 2016. “Synonymous Codons Direct Cotranslational Folding Toward Different Protein Conformations.” Molecular Cell 61: 341–351. 10.1016/j.molcel.2016.01.008 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
